Aim and Scope
Here we calculate the necessary data for the Island Biogeographic Analyses of the Caribbean Herpetofauna. We will be determining the species richness (SR) and phylogenetic species variability (PSV) for each island bank in the Caribbean. Each of these values are going to be calculated for various species groups (i.e. clades) as determined by the taxonomic hierarchies for the reptile and amphibian species present in the Caribbean Bioregion as delineated by Hedges et al. (2019) and including Bermuda.
The SR will also be calculated for clades that are delineated by using the phylogeny so that the group of species that are descendant from each node are a clade.
First we need to load in the caribmacro package.
# Project package
library(caribmacro)If you do not have this package installed, then run
devtools::load_all in order to load all of the necessary
functions if you are running this code in the caribmacro R project in
the shared folder. If you are not, then see the functions vignette for
the Functions needed.
# Load all of the functions of the caribmacro package
devtools::load_all("./", export_all = FALSE)
## Function needed ##
# 'hierarchy' -> returns the taxonomic hierarcy
# 'com_matrix' -> determines the communities within geographic features
# 'SR_geo' -> calculates the species richness within geographic features
# 'PSV_geo' -> calculates the psv within geographic features
# 'clade_age' -> calculates the crown age of clades
# 'node_clades' -> determines the clades of species based on phylogenetic nodes
# 'clade_area' -> determines the number of geographic feature each clade is found on
# 'row_max' -> determines the maximum value in each row of a data frame
# 'factorize' -> turns character columns of a data frame into factorsThe other R packages needed for this are:
## here() starts at C:/Users/tun71056/OneDrive - Temple University/Shared drives/Caribbean Macrosystem/caribmacro## Linking to GEOS 3.9.1, GDAL 3.4.3, PROJ 7.2.1; sf_use_s2() is TRUE## Loading required package: sp## rgeos version: 0.5-9, (SVN revision 684)
## GEOS runtime version: 3.9.1-CAPI-1.14.2
## Please note that rgeos will be retired by the end of 2023,
## plan transition to sf functions using GEOS at your earliest convenience.
## GEOS using OverlayNG
## Linking to sp version: 1.5-0
## Polygon checking: TRUEIsland Banks
For our analyses we will be clustering the islands in the Caribbean into banks based on Powell and Henderson 2012 and Hedges et al. 2019. To further delineate banks we used the General Bathymetric Chart of the Oceans (GEBCO Compilation Group 2020, downloaded September 2020) to group islands based on the under water topography.
Let’s now take a look at the banks.
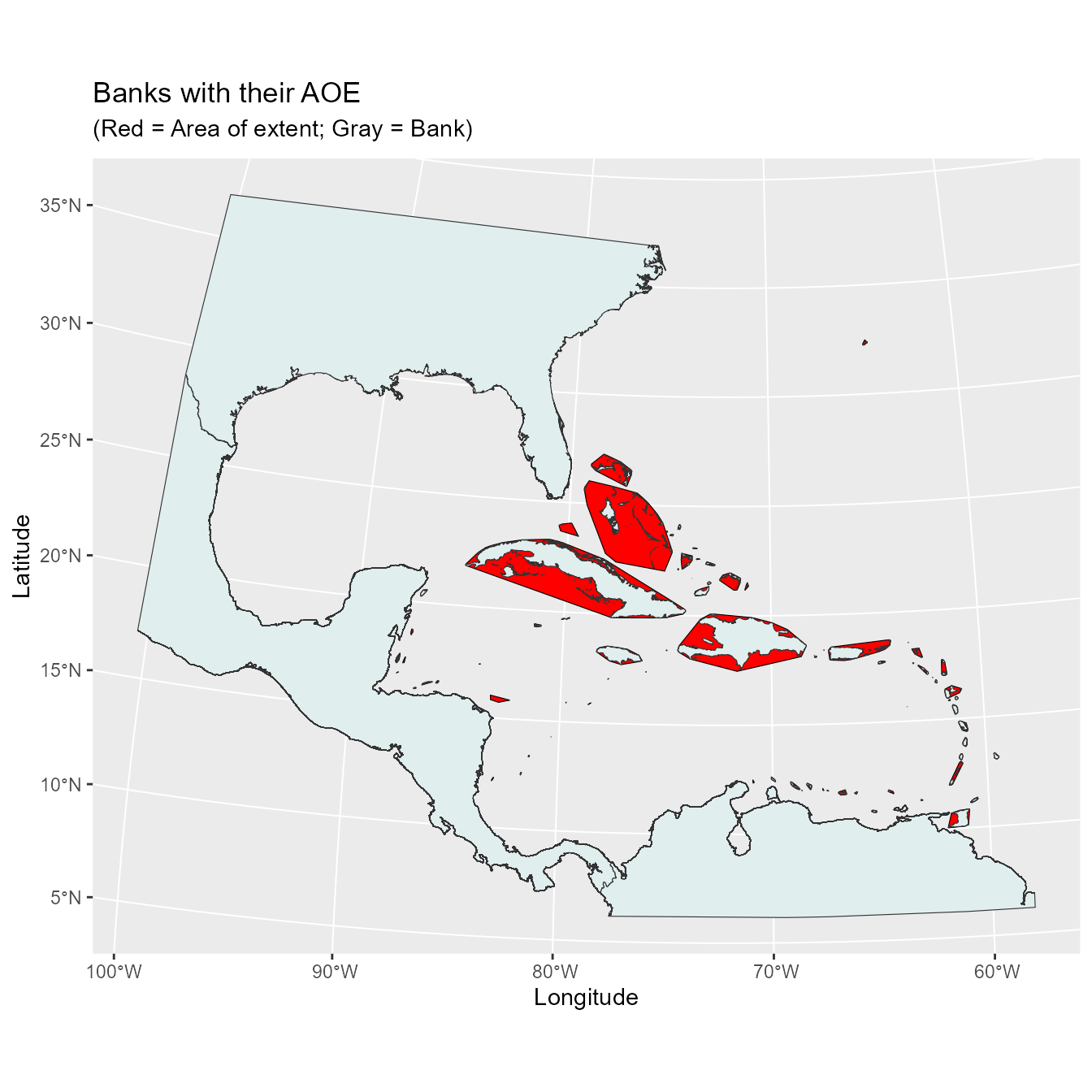
Taxonomic Clade Communities
Community Attributes
Now that we have determined bank area, we can now start working on the Community attributes. These attributes are their species richness (SR) and phylogenetic species variability (PSV).
Species Hierarchy
We will calculate SR and PSV for various clades of species on each
bank. Therefore, we need to determine the species hierarchies first. We
will do this using a function in the ‘caribherp’ package. This function
is hierarchy() and uses the package ‘taxize’ to look up and
download species hierarchies. The function in ‘taxize’ it uses makes
mistakes, and therefore, we need to check any errors outside of R. Once
any errors are fixed we then load the data back in. When we checked our
species hierarchies we saw that a couple species names were matched with
an insect or a plant species. The rest of the errors were all due to
misspellings or ids that were only to the genus level.
For this we need to read in the species occurrence data in the ‘data_raw’ folder.
herp <- read.csv(file.path(here::here(), 'data_raw', 'IBT_Herp_Records_final_SR_Drivers.csv'), header=TRUE)With the species occurrence data we can now run the
hierarchy() function and save the resulting data.
carib.hier <- hierarchy(data = herp, species = 'binomial', db = 'gbif')
#Need to check/correct any errors and missing hierarchies in Excel
write.csv(carib.hier, file.path(here(), 'data_raw', 'supporting_data', 'IBT_Carib_Sp_Taxonomy_v0.csv'), row.names=FALSE)Remember, we need to check the data outside R for errors and fix those errors. Once we do that we can then load in the new fixed hierarchies and merge it with the occurrence data.
carib.hier <- read.csv(file.path(here(), 'data_raw', 'supporting_data', 'IBT_Carib_Sp_Taxonomy_v1.csv'), header=TRUE)
# Combine Hierarchies with Occurrence Data
names(carib.hier)[which(names(carib.hier) == 'sp_in_Data')] <- 'binomial'
herp <- merge(herp, carib.hier, by = 'binomial', all = TRUE)Let’s check once more to see if we are missing any hierarchies
# Check for species missing their hierarchy
herp$binomial <- as.factor(herp$binomial)
chk <- levels(droplevels(herp[is.na(herp$Genus), 'binomial']))
chk## character(0)We can see that there are no missing hierarchies in our species
occurrence data. However, the hierarchy() function only
finds a species “typical” taxonomic hierarchy. If we want to add in
certain groups that are not a kingdom, phylum, class, order, family, or
genus, than we need to add them manually. Because we are interested in
all snakes, geckos, and skinks , we will include the species’ suborders
or smaller groups so that we include groups of interest. For instance,
the non-Iguania squamates were broken up into the infraorders Gekkota,
Amphisbaenia, Scincomorpha, and Anguimorpha (which combines the
infraorders Diploglossa and Platynota in our data) and the superfamily
Gymnophthalmoidea.
The suborder/infraorder groups were determined by searching the species’ families on the Integrated Taxonomic Information System (ITIS) and then doing a web search if the family’s ITIS standard report page did not list a suborder/infraorder.
However, the searches on ITIS illuminated a potential error in the identification of the Natricidae family. The family has is typically been considered a subfamily of Colubridae (Pryon et al. 2011). Therefore, we will change this family to match ITIS.
# Fix Family Error
herp[which(herp$Family=='Natricidae'), 'Family'] <- 'Colubridae'
# Add suborder groups
sorder <- read.csv(file.path(here(), 'data_raw', 'supporting_data', 'IBT_Carib_Suborders.csv'), header=TRUE)
herp <- merge(herp, sorder, by = 'Family', all.x = TRUE)
carib.hier <- merge(carib.hier, sorder, by = 'Family', all.x = TRUE)
chk <- levels(droplevels(as.factor(herp[is.na(herp$Suborder), 'Family'])))
chk## character(0)Community Matrices
Now that we have the hierarches for each species in our occurrence data, we can create the community matrices for each clade. These matrices will comprise of j columns, where j equals the number of species in the clade, and k rows, where k equals the number of banks the clade is recorded on. Each cell is then populated by a 1 or 0, where 1 represents the presence of the jth species on the kth bank, and 0 represents its absence.
In order to create these matrices we will use the
com_matrix() function from the ‘caribmacro’ package. This
function returns a named list of i community matrices where i is the
number of species groups. In our case we will create a community matrix
for all herpetofuana and all of the classes, orders, families, and
genuses in our occurrence data. Additionally, we will break up each of
these clades based on thier status on the banks (i.e. native or exotic)
and the entire clade. This comes out to be 693 species groups.
# First simplify bank status
herp$stat_new <- NA
for (i in 1:nrow(herp)) {
if (!is.na(herp[i, 'bnk_status'])) {
if (herp[i, 'bnk_status'] == 'E') {
herp[i, 'stat_new'] <- 'E'
} else if (herp[i, 'bnk_status'] == 'FE') {
herp[i, 'stat_new'] <- 'N'
} else if (herp[i, 'bnk_status'] == 'N') {
herp[i, 'stat_new'] <- 'N'
} else if (herp[i, 'bnk_status'] == 'PX') {
herp[i, 'stat_new'] <- 'N'
} else if (herp[i, 'bnk_status'] == 'U') {
herp[i, 'stat_new'] <- 'E'
} else {
herp[i, 'stat_new'] <- 'X'
}
} else {
herp[i, 'stat_new'] <- 'X'
}
}
# Fix specific statuses
## Considered extinct
herp[which(herp$binomial == 'Crocodylus rhombifer' & herp$bank == 'grand cayman'), 'stat_new'] <- 'X'
## Considered introduced by caribherp.org
herp[which(herp$binomial == 'Chelonoidis carbonarius' & herp$bank == 'anguilla'), 'stat_new'] <- 'E'
system.time(coms <- com_matrix(data = herp,
species = "binomial",
geo_group = "bank",
taxa_group = c("Genus", "Family", "Suborder", "Order", "Class"),
status = "stat_new",
stat_levels = c("N", "E"),
total = TRUE))## user system elapsed
## 6.19 0.15 6.34
# List the statuses excluded from the analyses
levels(as.factor(herp[which(herp$stat_new == 'X'), 'bnk_status']))## [1] "EI" "F" "FE" "G" "H" "W"For our analyses we are changing ‘unknown’ (U) statuses to ‘exotic’ (E) and the ‘possibly extinct’ (PE) and ‘have fossil but extant’ (FE) statuses to ‘native’ (N). We are also excluding ‘only known by fossils’ (F), ‘waifs’ (W), ‘extirpated exotics’ (EI), and records only identified to the genus level but the genus is found elsewhere on the bank (G).
Because our the way we are grouping species hierarchical, we may have clades that have the same species (e.g. the suborder clade Eusuchia has all of the same species as the order Crocodilia). Let’s check to see if this occurs in our data by looking at how many clades there are in the each level downstream from a clade. Those clades that only have one clade in a lower level will have the same species as the lower level.
tst <- herp
tst$binomial <- as.factor(tst$binomial)
grps <- c("Class", "Order", "Suborder", "Family", "Genus")
lists <- vector('list', length(grps))
names(lists) <- grps
for (i in 1:length(grps)) {
tst[, grps[i]] <- as.factor(tst[, grps[i]])
lists[[grps[i]]] <- levels(tst[, grps[i]])
}
out <- data.frame(NULL)
for (i in 1:length(lists)) {
if (i != length(lists)) {
for (j in 1:length(lists[[i]])) {
temp <- droplevels(tst[which(tst[, names(lists)[i]] == lists[[i]][j]),
c('binomial', grps[(i+1):length(grps)])])
for (k in 2:ncol(temp)) {
if (length(levels(temp[, k])) > 1) {
lvls <- levels(temp[, k])
tmp <- data.frame(Group.1 = rep(lists[[i]][j], length(lvls)),
Grp.1.lvl = rep(grps[i], length(lvls)),
Group.2 = lvls,
Grp.2.lvl = rep(which(grps == names(temp)[k]), length(lvls)),
Different = rep(TRUE, length(lvls)))
out <- rbind(out, tmp)
} else {
lvls <- levels(temp[, k])
tmp <- data.frame(Group.1 = rep(lists[[i]][j], length(lvls)),
Grp.1.lvl = rep(i, length(lvls)),
Group.2 = lvls,
Grp.2.lvl = rep(which(grps == names(temp)[k]), length(lvls)),
Different = rep(FALSE, length(lvls)))
out <- rbind(out, tmp)
}
}
}
} else {NA}
}
rm(tst, temp, tmp, lists, grps, lvls)
# To check that the code work:
# View(out[!out$Different, ])
rem <- levels(as.factor(out[!out$Different, 'Group.1']))
cat(paste0('Clades removed (n = ', length(rem), ') \n', '\n'), paste(rem, collapse = " | "))## Clades removed (n = 29)
##
## Agamidae | Amphisbaenidae | Anguidae | Anomalepididae | Cadeidae | Caudata | Centrolenidae | Crocodylia | Crocodylidae | Dactyloidae | Elapidae | Eleutherodactylidae | Eublepharidae | Geoemydidae | Hemiphractidae | Kinosternidae | Leiocephalidae | Leptodactylidae | Mesobatrachia | Pelomedusidae | Phrynosomatidae | Pipidae | Polychrotidae | Salamandridae | Salamandroidea | Strabomantidae | Tropidophiidae | Varanidae | XantusiidaeAs you can see there are a lot of clades that contain the same species as younger clades. Because this would be psuedoreplication (i.e. the same species group analyzed twice), we need to remove these clades from our analyses.
good <- gsub('.T', '', names(coms)[grep('.T', names(coms), fixed = TRUE)], fixed = TRUE)
good <- paste(good, ".", sep = "")
rem <- paste(rem, ".", sep = "")
good <- setdiff(good, rem)
tmp <- coms[names(coms)[grep(good[1], names(coms), fixed = TRUE)]]
for (i in 2:length(good)) {
tmp <- c(tmp, coms[names(coms)[grep(good[i], names(coms), fixed = TRUE)]])
}
coms <- tmp
rm(good, tmp, rem)Species Richness Calculation
Now that we have the community matrices we can calculate the species
richness (SR) of each species group on each bank. To do this we will use
the SR_geo() function in the ‘caribmacro’ package.
system.time(SR <- SR_geo(data = coms, geo_group = "bank"))## user system elapsed
## 2.29 0.02 2.31
SR_out <- SRClade Species Richness
We may also want to include the species richness of each clade in our analysis, because the clades may differ in their speciation rates. We will only calculate this for the Caribbean Bioregion. Therefore, the SR of each clade will represent the rate a clade will add a species due to speciation (for the native species richness) or both speciation and species introduction (both native and exotic species richness) in the Caribbean Bioregion.
We will calculate clade SR in a similar way to bank SR (see above).
However, instead of setting geo_group = "bank", we will set
geo_group = "region" in the com_matrix()
function, where ‘region’ is set to ‘carib’ representing the
Caribbean.
herp$region <- 'carib'
clade.coms <- com_matrix(data = herp,
species = "binomial",
geo_group = "region",
taxa_group = c("Genus", "Family", "Suborder", "Order", "Class"),
status = "stat_new",
stat_levels = c("N", "E"),
total = TRUE)
clade.SR <- SR_geo(data = clade.coms, geo_group = "region")
tmp <- strsplit(names(clade.SR)[which(names(clade.SR) != 'region')], ".", fixed = TRUE)
tmp <- data.frame(matrix(unlist(tmp), nrow = length(tmp), byrow = TRUE))
clade.SR <- cbind(tmp, as.numeric(clade.SR[1, names(clade.SR)[which(names(clade.SR) != 'region')]]))
names(clade.SR) <- c('Taxon', 'Group', 'SR')Clade P/A Matrix
It may be important to have a matrix that has the presence and absence of species in each clade
sp <- levels(as.factor(herp$binomial))
sp <- gsub(' ', '_', sp, fixed = TRUE)
cld.mat <- as.data.frame(matrix(NA, nrow(clade.SR), length(sp)))
names(cld.mat) <- sp
cld.mat$Clade <- clade.SR$Taxon
cld.mat$Group <- clade.SR$Group
cld.mat <- cld.mat[, c('Clade', 'Group', sort(names(cld.mat)[which(names(cld.mat) != 'Clade' &
names(cld.mat) != 'Group')]))]
for (i in 1:nrow(cld.mat)) {
if (cld.mat[i, 'Group'] != 'T') {
if (is.element(cld.mat[i, 'Clade'], herp$Class)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Class == cld.mat[i, 'Clade'] & herp$stat_new == cld.mat[i, 'Group']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Order)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Order == cld.mat[i, 'Clade'] & herp$stat_new == cld.mat[i, 'Group']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff( names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Suborder)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Suborder == cld.mat[i, 'Clade'] & herp$stat_new == cld.mat[i, 'Group']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Family)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Family == cld.mat[i, 'Clade'] & herp$stat_new == cld.mat[i, 'Group']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Genus)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Genus == cld.mat[i, 'Clade'] & herp$stat_new == cld.mat[i, 'Group']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else {NA}
} else {
if (is.element(cld.mat[i, 'Clade'], herp$Class)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Class == cld.mat[i, 'Clade']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Order)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Order == cld.mat[i, 'Clade']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Suborder)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Suborder == cld.mat[i, 'Clade']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Family)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Family == cld.mat[i, 'Clade']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else if (is.element(cld.mat[i, 'Clade'], herp$Genus)) {
tmp.sp <- levels(as.factor(droplevels(herp[which(herp$Genus == cld.mat[i, 'Clade']), 'binomial'])))
tmp.sp <- gsub(' ', '_', tmp.sp, fixed = TRUE)
cld.mat[i, tmp.sp] <- 1
cld.mat[i, setdiff(names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')], tmp.sp)] <- 0
} else {NA}
}
}
cld.mat[which(cld.mat$Clade == 'All'), names(cld.mat)[which(names(cld.mat) != 'Clade' & names(cld.mat) != 'Group')]] <- 1Data Saving
Now lets save all of the community and clade attributes we calculated in the ‘data_out’ folder. We also want to save the clade SR and the Caribbean species taxonomy in the supp_info folder.
# Data for data_out
write.csv(SR_out, file.path(here(), 'data_out', 'sr_drivers', 'SR_Drivers_Herp_SR.csv'), row.names = FALSE)
write.csv(clade.SR, file.path(here(), 'data_out', 'sr_drivers', 'SR_Drivers_Clade_SR.csv'), row.names = FALSE)
# Data for supp_info
write.csv(carib.hier, file.path(here(), 'data_out', 'sr_drivers', 'supp_info', 'SR_Drivers_Carib_Taxonomy.csv'), row.names=FALSE)
write.csv(cld.mat, file.path(here(), 'data_out', 'sr_drivers', 'supp_info', 'SR_Drivers_Taxonomy_Clade_Matrix.csv'), row.names=FALSE)Temple University, jmg5214@gmail.com↩︎