Species Distribution Modeling
vignette-011-suitability-models.RmdHow to Build, Evaluate, and Save Species Distribution Models for Ailanthus altissima (TOH) and Lycorma delicatula (SLF)
This vignette is designed to demonstrate the workflow that lead to
the creation of the three species distribution models (SDMs) used to
assess the risk of SLF paninvasion. The description
will not include actual running of some steps of this workflow, as some
are unwieldy. Instead, information to ensure proper replication of those
steps is provided. Notably, the MaxEnt runs are not conducted in this
vignette, although the means to do so within R may be
possible.
The scope of this vignette is as follows:
- Reading and Visualization of Presence Data
- Preparation of Spatial Data
- Evaluation of Spatial Data Collinearity
- Building and evaluating MaxEnt SDMs
- Visualizing SDMs and Extracting Summary Statistics by Geopolitical Unit
Setup
We load the packages that are necessary to complete any analyses that allow us to demonstrate the workflow. Packages may include comments to clarify their purpose or the steps in which they are used.
library(slfrsk) #this package, has extract_enm()
library(tidyverse) #data manipulation
library(here) #making directory pathways easier on different instances
library(ENMTools) #enviro collinearity analyses
library(patchwork) #easy combined plots
library(scales) #rescale the plots easily
library(rgdal) #load shapefiles
library(doParallel) #parallized extraction1. Reading and Visualization of Presence Data
We built models of both SLF and TOH from presence records obtained from public databases (as seen in the “GBIF Records” vignette). As is standard practice, we checked all records for quality, removed duplicate and imprecise records, and obtained 8,022 TOH and 325 SLF unique and cleaned presence records.
#load data
data("slf_points")
data("toh_points")
#plot points on map: TOH
map_toh <- ggplot() +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = "#FFFFFF", color = "black", lwd = 0.15) + #world map
geom_point(data = toh_points, aes(x = x, y = y), color = "#0072b2", alpha = 0.10) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
#plot.background = element_rect(fill ="#f0f8ff"),
panel.background = element_rect(fill = "#f0f8ff")) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "TOH Presence")
#plot points on map: SLF
map_slf <- ggplot() +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = "#FFFFFF", color = "black", lwd = 0.15) + #world map
geom_point(data = slf_points, aes(x = x, y = y), color = "#d55e00", alpha = 0.50) +
theme_bw() +
labs(x = "", y = "") +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
#plot.background = element_rect(fill ="#f0f8ff"),
panel.background = element_rect(fill = "#f0f8ff")) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "SLF Presence")
#patchwork output of both maps
map_toh / map_slf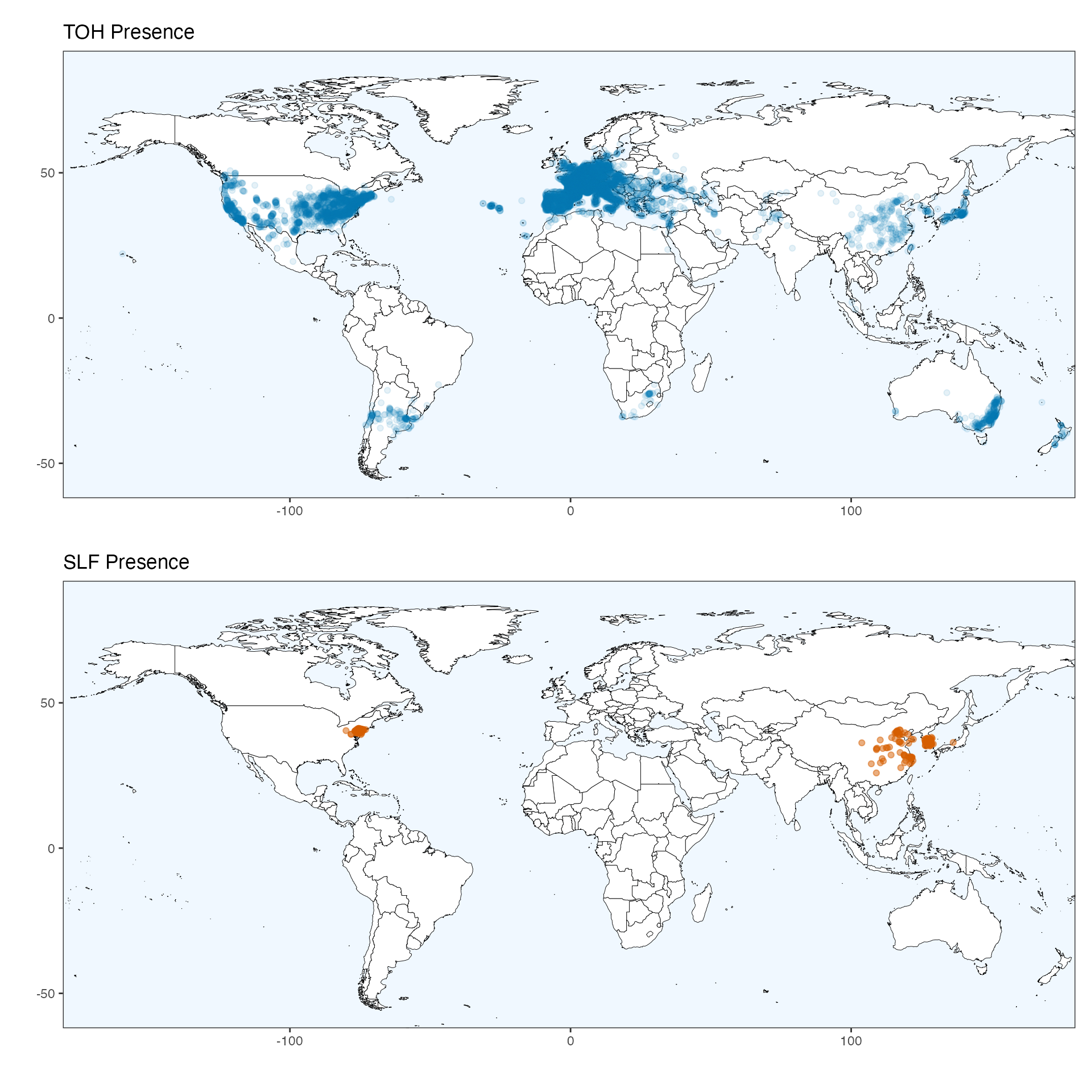
#ggsave((map_toh / map_slf), filename = file.path(here(), "slf_toh_presence.png"), height = 10, width = 10)
ggplot() +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = "#FFFFFF", color = "black", lwd = 0.15) + #world map
geom_point(data = toh_points, aes(x = x, y = y), color = "#0072b2", alpha = 0.10) +
geom_point(data = slf_points, aes(x = x, y = y), color = "#d55e00", alpha = 0.50) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
#plot.background = element_rect(fill ="#f0f8ff"),
panel.background = element_rect(fill = "#f0f8ff")) +
#theme_bw() +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85))2. Preparation of Spatial Data
To discern which environmental variables to use in the species distribution models (SDMs), we had to minimize collinearity among included covariates (A. Townsend Peterson et al. (2011)). To do so, we estimated pairwise correlation coefficients for 22 global raster covariates hypothesized to influence SLF and TOH distributions: the 20 WorldClim topographic and bioclimatic variables (Hijmans et al. (2005), Fick and Hijmans (2017)), forest height (Simard et al. (2011)), and access to cities (Weiss et al. (2018)).
To limit the raster data of covariates to the area of interest, we
cropped their extent to \([-180, 180]\)
for latitude and \([-60, 84]\) for
longitude and ensured that the extent and resolutions matched one
another by using one layer as a reference raster with
raster::resample(method = "bilinear). This corrected any
mismatching among cropped rasters of different sources (example code
below).
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#ensure that extent is identical
#get file names
env.files <- list.files(file.path(mypath, "originals"), pattern = "[.]tif", full.names = T)
env.short <- list.files(file.path(mypath, "originals"), pattern = "[.]tif", full.names = F)
#change the labeling of the output layers
output.files <- env.short
#change weird BIOCLIM prefix
output.files <- gsub(pattern = "wc2.0_bio_30s", replacement = "global_bio", x = output.files)
#change weird ENVIREM prefix
output.files <- gsub(pattern = "current_30arcsec", replacement = "global_env", x = output.files)
#change weird ATC prefix
output.files <- gsub(pattern = "2015_accessibility_to_cities_v1.0", replacement = "global_atc", x = output.files)
#crop one of the BIOCLIM layers to set the bounding box and resolution to have files fixed
same.extent <- extent(-180, 180, -60, 84)
main_layer <- crop(raster(file.path(mypath,"originals", "wc2.0_bio_30s_01.tif")), y = same.extent, overwrite = F)
#reset extent stepwise
for(a in seq_along(env.files)){
#ensure that the CRS is consistent
rast.hold <- raster(env.files[a])
crs(rast.hold) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
#resample to fit the extent/resolution of the reference BIOCLIM layer
#use bilinear interpolation, since values are continuous
rast.hold <- resample(x = rast.hold, y = main_layer, method = "bilinear")
#write out the new resampled rasters!
writeRaster(x = rast.hold, filename = file.path(mypath,"v1", output.files[a]), overwrite = T)
}
} #closes no run ifFor tractability, we first downsized the resolution of these global
covariate rasters with
raster::aggregate(fact = 2, fun = mean, expand = TRUE, na.rm = TRUE)
before assessing collinearity (see example code below). However, the
full resolution covariates were used to build all models.
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#list the enviro layers to load sequentially
env.files <- list.files(path = file.path(mypath, "v1"), pattern = ".tif", full.names = T)
env.short <- list.files(path = file.path(mypath, "v1"), pattern = ".tif", full.names = F)
#downsampling by a factor of 2 (read 2 cells deep around a cell) and take the mean of the cells
for(a in seq_along(env.files)){
holder <- raster(env.files[a])
down_holder <- raster::aggregate(holder, fact = 2, fun = mean, expand = TRUE, na.rm = TRUE, filename =file.path(mypath, "v1_downsampled", env.short[a]), overwrite = T)
}
} #closes no run if3. Evaluation of Spatial Data Collinearity
To evaluate the correlations among the covariates, we used
ENMTools::raster.cor.matrix(method = "pearson"). Note that
this code may still take quite some time to run. For brevity, we have
included a version of the correlation matrix with the compendium for
quick review (location:
/data-raw/env_cor_v1_downsampled.csv).
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#load the downsampled layers and stack for raster.cor.matrix command
#list of layer paths
env.files <- list.files(path = file.path(mypath,"v1_downsampled"), pattern = ".tif", full.names = T)
#stack the downsized layers
env <- raster::stack(env.files)
#evaluate correlations for raster layers
#create a correlation matrix for picking model layers
env.corr <- ENMTools::raster.cor.matrix(env, method = "pearson")
#write out the correlations as a csv
write.csv(x = env.corr, file = file.path(here(),"data-raw", "env_cor_v1_downsampled.csv"), col.names = TRUE, row.names = TRUE)
} #closes no run ifTo visualize the correlations between pairs of environmental covariates easily, we take the absolute value of all correlations and present the strength of relationships as a heatmap.
#re-read the correlation table in again
env.cor <- read.csv(file = file.path(here::here(),"data-raw", "env_cor_v1_downsampled.csv"), row.names = 1)
#here, we make cor's absolute values and make the data tidy to make it easier to plot in ggplot2
p_env_cor <- env.cor %>%
abs(.) %>%
as_tibble() %>%
mutate(covar = colnames(.)) %>%
dplyr::select(covar, everything()) %>%
pivot_longer(cols = -covar, names_to = "var") %>%
dplyr::select(var, covar, everything()) %>%
ggplot() +
geom_tile(aes(x = var, y = covar, fill = value)) +
viridis::scale_fill_viridis(discrete = FALSE, direction = -1, limits = c(0,1), name = "abs(Correlation)") +
guides(x = guide_axis(angle = 90)) +
labs(x = "", y = "")
p_env_cor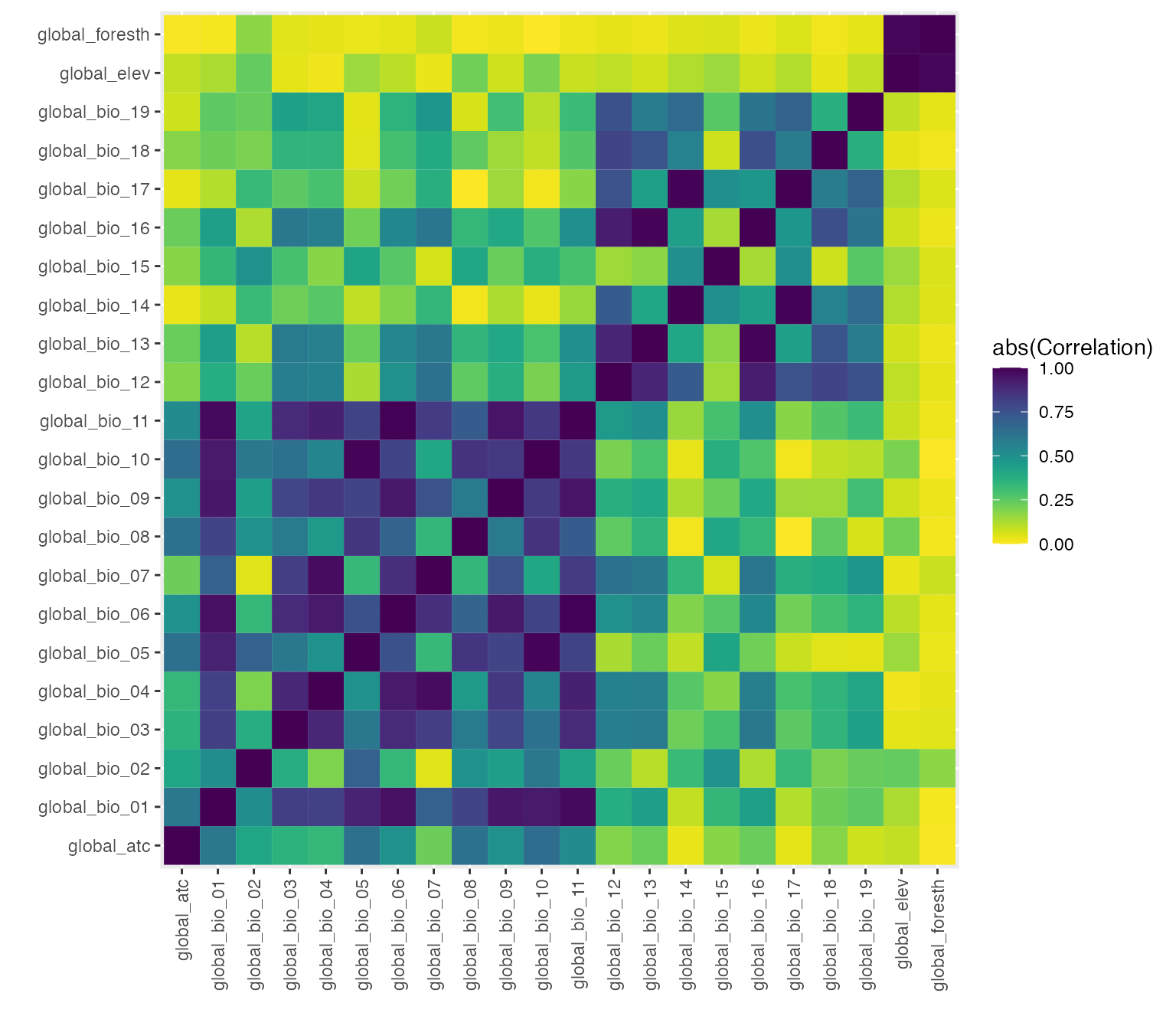
#ggsave(p_env_cor, filename = file.path(here(), "p_env_cor.png"), height = 10, width = 10)With the correlations, we identified 6 of the 22 covariates that had
minimal cross-correlations: annual mean temperature
(BIO01), mean diurnal temperature range
(BIO02), annual precipitation (BIO12),
precipitation seasonality (BIO15), elevation
(ELEV), and access to cities (ATC).
These variables were then converted to ASCII files (.asc),
as required by MaxEnt (example code below). We also set all
NA values to -9999, which is the default value
recognized as NA by MaxEnt.
Note that these .asc files are massive. It is not
advisable to run this chunk unless you are certain that you have
sufficient space to do so. It is for this reason (in part) that we do
not provide this file type for all covariates.
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#get and set file names
env.files <- list.files(path = file.path(mypath,"v1"), pattern = "[.]tif", full.names = T)
env.short <- list.files(path = file.path(mypath,"v1"), pattern = "[.]tif", full.names = F)
env.asc <- gsub(pattern = ".tif", replacement = ".asc", x = env.short)
#loop to convert and make sure to set NA values to -9999
for(a in seq_along(env.files)){
file_to_asc <- raster(env.files[a])
NAvalue(file_to_asc) <- -9999
writeRaster(x = file_to_asc, filename = file.path(mypath, "v1_maxent", env.asc[a]), format = "ascii", overwrite = F)
}
} #closes no run if4. Building and evaluating MaxEnt SDMs
Focal Models
To further evaluate environmental covariates, we modeled each of them individually with SLF. To do this, we used MaxEnt (v3.4.1, available at https://biodiversityinformatics.amnh.org/open_source/maxent/) under default parameters, excluding the following changes (Phillips, Anderson, and Schapire (2006), Pearson et al. (2007)):
- All feature types were made available but still set to
Auto Features(Linear, Quadratic, Product, Threshold, and Hinge set toTRUEbefore setting Auto Features toTRUE) -
Create response curveswas set toTRUE -
Do jackknife to measure variable importancewas set toTRUE -
Replicatesset to5for SLF (this sets the number of k-fold crossvalidation replicates and determines the test proportion from k) -
Apply threshold ruleset toMinimum training presence -
Threadsset to the available number of processor threads available
After fitting individual models, we fit these 6 covariates combined
to SLF and TOH presences, except that the number of
Replicates = 10 for TOH, rather than
Replicates = 5. The resultant two models are:
- sdm_toh—a multivariate SDM of TOH
- sdm_slf1—a multivariate SDM of SLF
A third model was created from sdm_toh suitability and SLF presence records, based on evidence supporting the strong affinity of SLF for TOH as a preferred host (Parra, Moylett, and Bulluck (2018), (urban_perspective:_2019?)):
- sdm_slf2—a single variate SDM of SLF
Here, we report in a simple table the metrics used to evaluate the various models that were considered as well as the percent contribution of each variable to each model (where applicable). Model validation was conducted with k-fold cross-validation (k partitions discussed above) via evaluation of the receiver operating characteristic of the AUC and omission error (Fielding and Bell (1997), Phillips, Anderson, and Schapire (2006), Pearson et al. (2007), Anderson and Gonzalez (2011)).
For AUC, the fraction of true positives relative to type I error (positive background points) is compared at all possible thresholds for each model (Fielding and Bell (1997), Phillips, Anderson, and Schapire (2006)). The resultant plot’s area under the curve is assessed relative to a random model (\(AUC = 0.50\)), such that values close to \(1.00\) indicate strong model performance and those \(\leq 0.50\) suggest poor performance (Fielding and Bell (1997)). Given presence only data, measured AUC cannot reach \(1.00\), but model AUCs that approach \(1.00\) are considered to perform well (Wiley et al. (2003), Phillips, Anderson, and Schapire (2006)).
Given recent concerns with model evaluation with AUC (A. Townsend Peterson, Papeş, and Soberón (2008), Lobo, Jiménez-Valverde, and Real (2008), Jiménez‐Valverde (2012)), we confirmed model performance with average omission error, which measures the proportion of presence point(s) predicted with suitability less than the minimum training presence threshold (Pearson et al. (2007), Anderson and Gonzalez (2011)).
| model | avg_test_auc | avg_omission_error | atc_contribution | bio01_contribution | bio02_contribution | bio12_contribution | bio15_contribution | elev_contribution |
|---|---|---|---|---|---|---|---|---|
| sdm_toh | 0.7779 | 0.0003 | 61.03 | 36.74 | 0.06 | 1.08 | 0.81 | 0.28 |
| sdm_slf1 | 0.9828 | 0.0064 | 57.16 | 22.97 | 1.95 | 11.63 | 5.96 | 0.32 |
| sdm_slf2 | 0.9675 | 0.0032 | NA | NA | NA | NA | NA | NA |
The three models performed similarly, albeit with small differences across model performance metrics. Notably, the variables that contributed most the sdm_toh also contributed most to sdm_slf1, albeit at differing amounts. Thus, we created a consensus output map by averaging the three models as an ensemble.
To visualize the models and produce the ensemble model image, we
first had to convert the mean model for each set of replicates
(species_name_avg.asc) and convert it to a GeoTIFF
(.tif), which is more easily visualized and stored (example
code below).
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#read in file as ASCII raster
enm_data_slf <- raster(file.path(mypath, "maxent_models", "10_21_20_maxent_slf_full", "Lycorma_delicatula__White,_1845__avg.asc"))
#make sure CRS is WGS84
crs(enm_data_slf) <- "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0"
#write out as a geotiff
writeRaster(x = enm_data_slf, filename = file.path(mypath,"maxent_models", "slf.tif"), format = "GTiff")
} #closes no run ifEnsemble Model
To create the ensemble model image, we averaged the three models (see code below). The resultant model image was used to extract summary statistics by geopolitical unit.
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#read in a geotiff version of the maxent suitability for all three models
toh <- raster(file.path(mypath,"maxent_models", "toh.tif"))
slf <- raster(file.path(mypath,"maxent_models", "slf.tif"))
slftoh <- raster(file.path(mypath,"maxent_models", "slftoh.tif"))
#stack the rasters
enm_data <- stack(c(toh, slf, slftoh))
#make mean raster
enm_ensemble <- mean(enm_data)
#write out the resulting file
writeRaster(x = enm_ensemble, filename = file.path(mypath, "maxent_models", "slftoh_ensemble_mean.tif", format = "GTiff")
} #closes no run if5. Visualizing SDMs and Extracting Summary Statistics by Geopolitical Unit
Visualization
To make visualization more tractable, we reduced the size of the SDMs
by using
raster::aggregate(fun = mean, expand = TRUE, na.rm = TRUE)
(setting fact= to 4 for states
and 10 for countries) to reduce the SDM raster
resolution (more specific example code can be found in
data-raw/downsample_raster_models.R for downsampling and
data-raw/convert_data_rda.R for fortify()-ing
rasters to plot rasters easily with ggplot2). We also
clipped the SDMs to the continental US to produce the files used for
separate states and countries visualization.
The three models can be visualized below for the US and then the
world:
USA Maps
#read in the data
data("slf_usa_df")
data("slftoh_usa_df")
data("toh_usa_df")
#plot just the states and suitability first
sdm_toh_usa <- ggplot() +
geom_raster(data = toh_usa_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.9, show.legend = F) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('state'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.10) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-124.7628, -66.94889), ylim = c(24.52042, 49.3833)) +
ggtitle(label = "sdm_toh USA")
sdm_slf1_usa <- ggplot() +
geom_raster(data = slf_usa_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.9, show.legend = F) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('state'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.10) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-124.7628, -66.94889), ylim = c(24.52042, 49.3833)) +
ggtitle(label = "sdm_slf1 USA")
sdm_slf2_usa <- ggplot() +
geom_raster(data = slftoh_usa_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.9, show.legend = T) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('state'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.10) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-124.7628, -66.94889), ylim = c(24.52042, 49.3833)) +
ggtitle(label = "sdm_slf2 USA")
#visualize together
sdm_toh_usa / sdm_slf1_usa / sdm_slf2_usa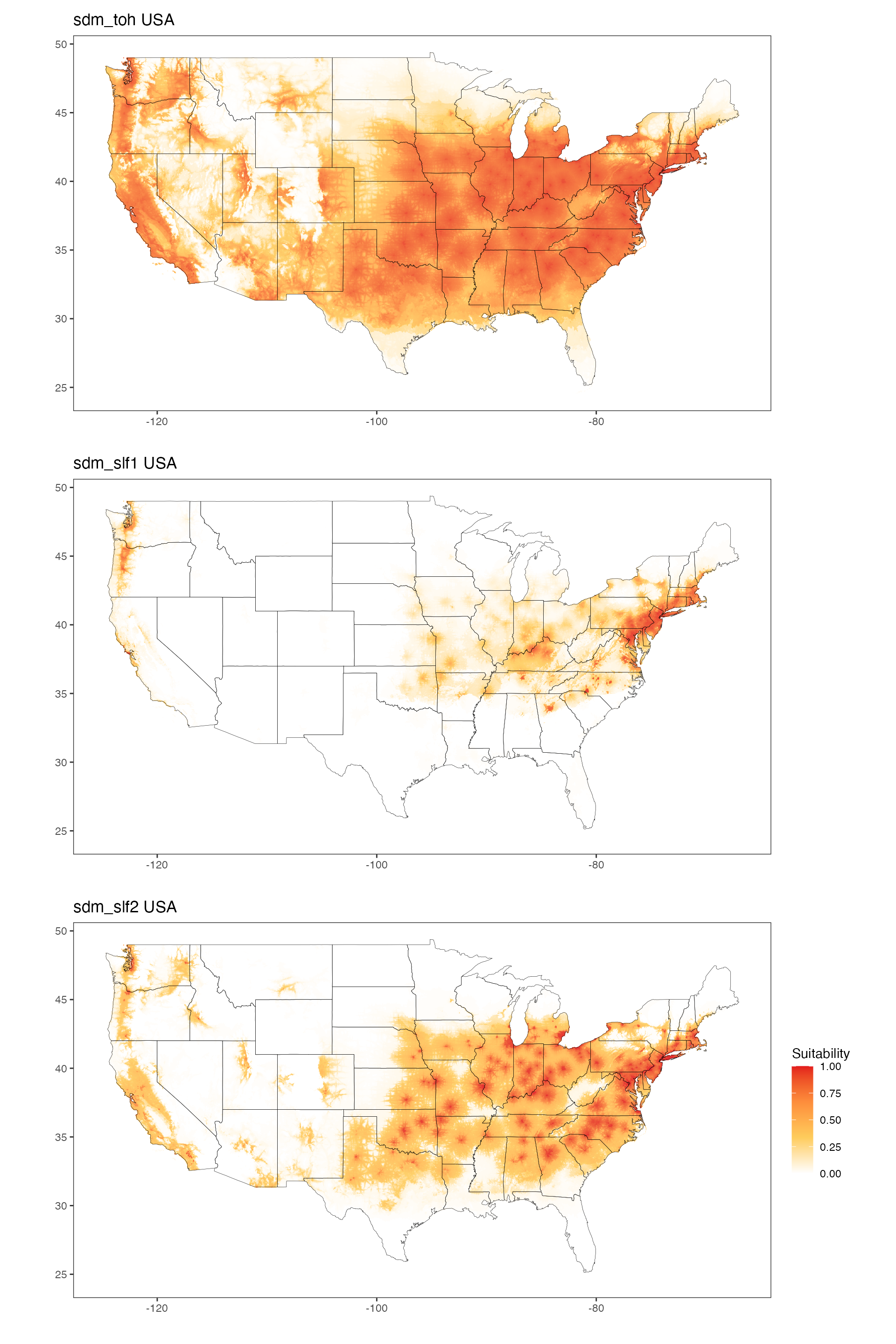
#ggsave(sdm_toh_usa, filename = file.path(here(), "sdm_toh_usa.png"), height = 5, width = 10)
#ggsave(sdm_slf1_usa, filename = file.path(here(), "sdm_slf1_usa.png"), height = 5, width = 10)
#ggsave(sdm_slf2_usa, filename = file.path(here(), "sdm_slf2_usa.png"), height = 5, width = 10)World Maps
Please note that the images for countries are downsized considerably to ensure that the visualization data can be included with the package.
#read in the data
data("slf_df")
data("slftoh_df")
data("toh_df")
#plot just the states and suitability first
sdm_toh <- ggplot() +
geom_raster(data = toh_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.8, show.legend = F) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.15) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "sdm_toh")
sdm_slf1 <- ggplot() +
geom_raster(data = slf_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.8, show.legend = F) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.15) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "sdm_slf1")
sdm_slf2 <- ggplot() +
geom_raster(data = slftoh_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.8, show.legend = T) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.15) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "sdm_slf2")
#visualize together
sdm_toh / sdm_slf1 / sdm_slf2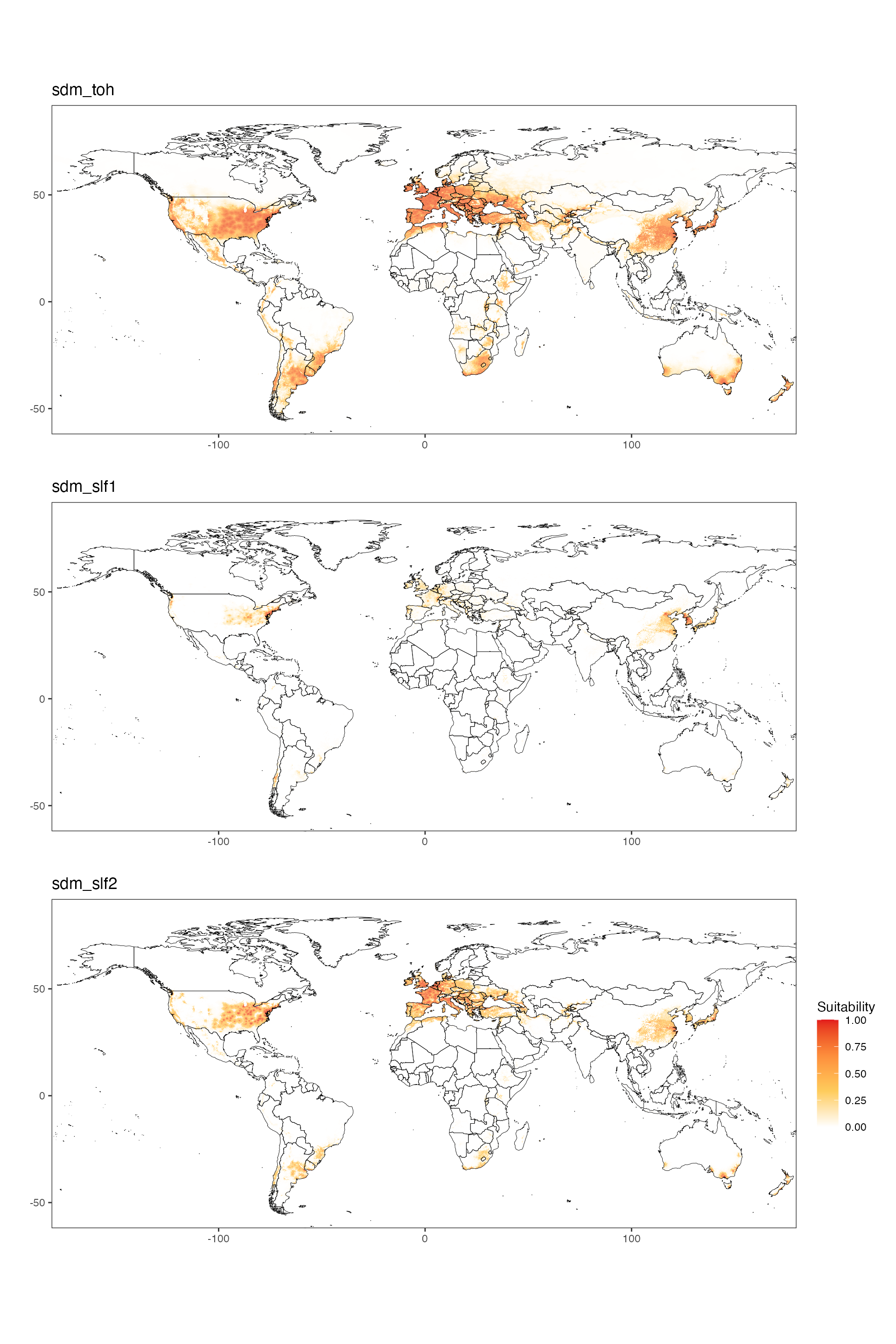
#ggsave(sdm_toh, filename = file.path(here(), "sdm_toh.png"), height = 5, width = 10)
#ggsave(sdm_slf1, filename = file.path(here(), "sdm_slf1.png"), height = 5, width = 10)
#ggsave(sdm_slf2, filename = file.path(here(), "sdm_slf2.png"), height = 5, width = 10)Ensemble Maps
We can also visualize the ensemble images similarly (see the
vignette-030-risk-maps for more detail versions). Note that
these visualizations are also downsampled, albeit at fact=4
for both maps.
#read in data
data("suitability_usa_df")
data("suitability_countries_df")
en_usa <- ggplot() +
geom_raster(data = suitability_usa_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.9, show.legend = F) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('state'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.10) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-124.7628, -66.94889), ylim = c(24.52042, 49.3833)) +
ggtitle(label = "USA Ensemble")
en <- ggplot() +
geom_raster(data = suitability_countries_df, aes(x = x, y = y, fill = rescale(value)), alpha=0.8, show.legend = T) +
scale_fill_gradientn(limits= c(0,1), name = "Suitability", colors = rev(c("#e31a1c","#fd8d3c", "#fecc5c", "#FFFFFF"))) +
geom_polygon(data = map_data('world'), aes(x = long, y = lat, group = group), fill = NA, color = "black", lwd = 0.15) +
theme_bw() +
labs(x = "", y = "") +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank()) +
coord_quickmap(xlim = c(-164.5, 163.5), ylim = c(-55,85)) +
ggtitle(label = "Ensemble")
#visualize together
en_usa / en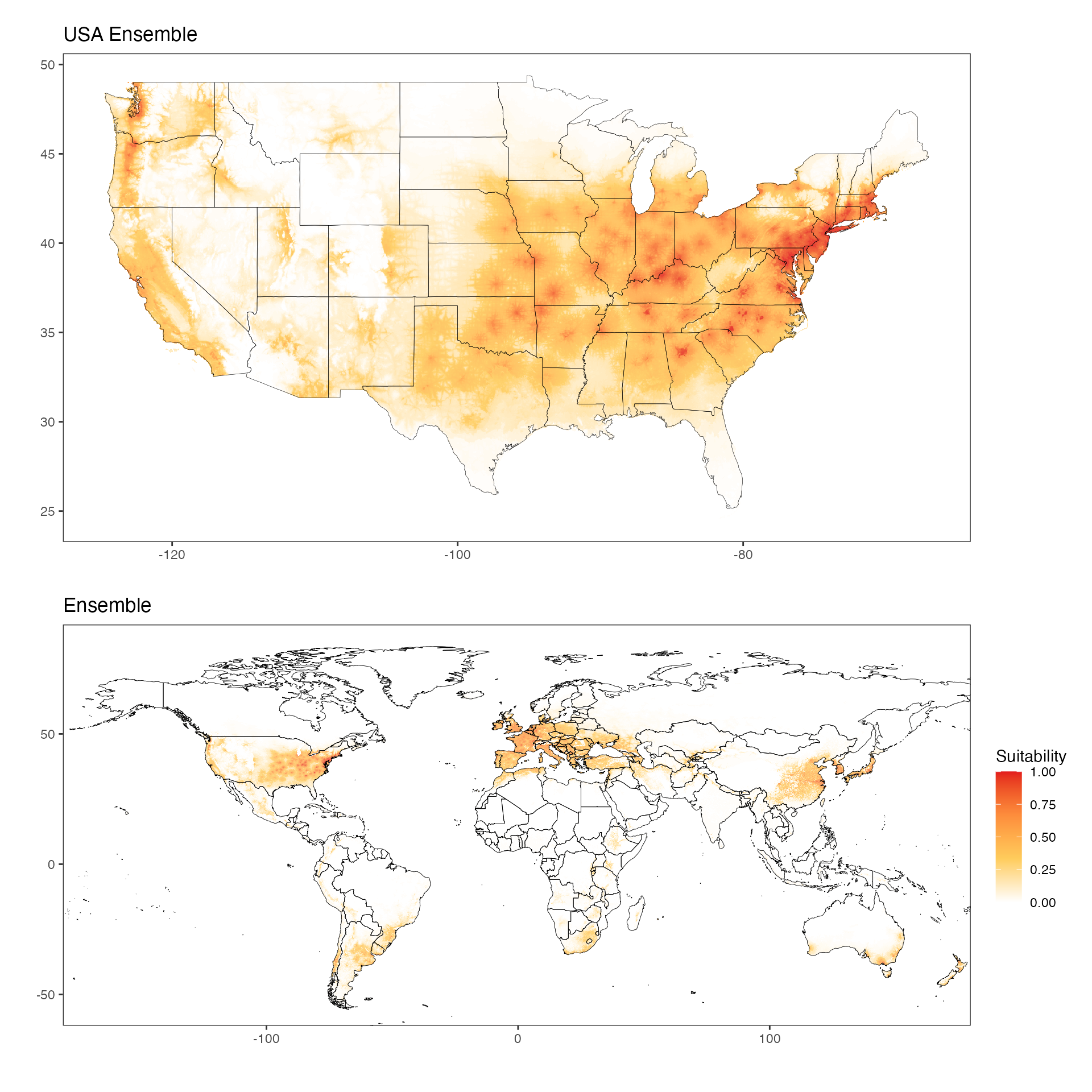
#ggsave(en_usa, filename = file.path(here(), "en_usa.png"), height = 5, width = 10)
#ggsave(en, filename = file.path(here(), "en.png"), height = 5, width = 10)Extraction of Summary Statistics
With the three original and ensemble SDMs, we extracted summary
statistics by geopolitical unit to later use for evaluation of
suitability. To do this, we use a custom function
(slfrsk::extract_enm()) that uses a reference shapefile (we
use a saved copy of GADM data v3.6 for the world and US—both can be
obtained online, https://gadm.org/data.html) to isolate the portion of
the raster contained within each geopolitical unit and calculate the
following summary statistics for suitability values:
- mean
- standard deviation
- minimum
- maximum
- quantiles at the following probabilities:
- 0.25
- 0.50 (median)
- 0.75
- 0.90
Here, we demonstrate how we used this function to extract the summary
statistics for each state and country used as
our measure of establishment potential. Note that we do
not use the save.plots argument or save the output. We also
use the .tif versions of full-scale models for these
extractions, due to their smaller size. We show code that works for the
ensemble but also include commented-out code to run for an
individual model and calculate rsa.
if(FALSE){ #open no run if
#my path to the GoogleDrive shared directory
mypath <- "/Volumes/GoogleDrive/Shared drives/slfData/data/slfrsk/raw_env"
#shapefile of world by countries
world <- readOGR(file.path(mypath, "geo_shapefiles", "gadm36_levels_shp", "gadm36_0.shp"), verbose = F, p4s = '+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0')
#shapefile of US states (same sourcce as usa data)
states <- readOGR(dsn = file.path(mypath, "geo_shapefiles", "gadm36_USA_shp", "gadm36_USA_1.shp"), verbose = F, p4s = '+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0')
#load data to test
#ensemble model
enm_ensemble <- raster(file.path(mypath, "maxent_models", "slftoh_ensemble_mean.tif"))
#example of running the function
#extract the states
states_extracts_ensemble <- extract_enm(enm = enm_ensemble, geoshape = states, id0 = "NAME_1", id = as.character(states$NAME_1))
#optional save the summary data
write_csv(path = file.path(here(),"data-raw", "extract_states_slftoh_ensemble_mean.csv"), x = states_extracts_ensemble[[1]], col_names = T)
#countries version
countries_extracts_ensemble <- extract_enm(enm = enm_ensemble, geoshape = world, id0 = "NAME_0", id = as.character(world$NAME_0))
write_csv(path = file.path(here(),"data-raw", "extract_countries_slftoh_ensemble_mean.csv"), x = countries_extracts_ensemble[[1]], col_names = T)
#parallelized version
countries_extracts_ensemble2 <- extract_enm2(enm = enm_ensemble, geoshape = world, id0 = "NAME_0", id = as.character(world$NAME_0), multipar = TRUE, ncores = 4)
#write out
write_csv(path = file.path(here(),"data-raw", "extract_countries_slftoh_ensemble_mean2.csv"), x = countries_extracts_ensemble2, col_names = T)
#example with s3
#the threshold if desired: enm_slftoh$`Minimum training presence Cloglog threshold`[nrow(enm_slftoh)]
#enm_data_slftoh <- raster(file.path(mypath, "maxent_models/11_07_18_maxent_slf+toh_+atc-bio02/lycorma_delicatula_avg.asc"))
#enm_data_slftoh <- raster(file.path(mypath, "maxent_models/slftoh.tif"))
#enm_slftoh <- read_csv(file.path(mypath, "maxent_models/11_07_18_maxent_slf+toh_+atc-bio02/maxentResults.csv"))
#state_extracts_slftoh <- extract_enm(enm = enm_data_slftoh, geoshape = states, id0 = "NAME_1", id = as.character(states$NAME_1), th = enm_slftoh$`Minimum training presence Cloglog threshold`[nrow(enm_slftoh)])
} #closes no run ifUltimately, the saved versions of these extracted suitabilities are
saved in /data-raw and then can be handled by the script in
the same directory, convert_data-rda.R, to be converted
into .rda file(s), which are stored in /data
and used by the other vignettes (with few, if any, exceptions that use a
shared GoogleDrive). Any exceptions should be reproducible with the
relevant raw starting files (INSERT HERE).