
Create risk maps of each model scale and intersect predictions
Samuel M. Owens1
2025-07-31
Source:vignettes/120_create_suitability_maps.Rmd
120_create_suitability_maps.RmdOverview
In the last vignette, I applied a weighting factor to the three regional-scale models to create an ensembled mean prediction. I have already created a global-scale model prediction, so now that the regional-scale models have been ensembled, I can directly compare the predictions made by the two modeled scales. Each model in this multi-scale analysis has been projected to the entire globe, so I can now measure the differences between these two models. My goals for application of these models are to look at the predictions for SLF risk of establishment made (A) spatially for globally important winegrowing countries, (B) at the specific locations of globally important viticultural regions, and (C) at the location of known SLF populations. I plan to explore the suitability for SLF via creating a range of outputs:
- risk maps
- range shift maps
- viticultural risk quadrant plots
- viticultural risk tables
- variable response curves
- other summary statistics, including AUC and variable contribution
In this vignette, I will create all mapping outputs to depict the risk of SLF establishment both historically and under future climate scenarios. I will also create a range shift map to show the expansion and contraction of suitable area under climate change. I will create these maps by intersecting the global and regional_ensemble model predictions to create classified (binarized) rasters. The level of agreement between each model prediction will be used to infer the level of SLF establishment risk.
The process will follow this general flow for creating the risk maps:
- Create suitable vs unsuitable classes based on the MTSS threshold of both rasters
- Convert each model raster to binary (suitable/unsuitable) based on these suitability thresholds
- Mosaic the two rasters and sum their values.
- Re-classify summed raster as areas where both agree on presence, both agree on absence, or one classifies and the other classifies absent (4 total categories)
I created a consensus MTSS threshold for each model in the previous vignette, which I will use to binarize these rasters. This procedure will be repeated for each ssp scenario and then a mean of the scenarios to produce different predictions based on different climate scenarios. I will also calculate the total area per risk category and raster to show how the change in suitable area might change depending on the ssp scenario.
Finally, I will plot a range shift map to show the expansion and contraction of suitable area per ssp scenario (essentially, the difference between the historical and future predictions).
Setup
# general tools
library(tidyverse) #data manipulation
library(here) #making directory pathways easier on different instances
# here() starts at the root folder of this package.
library(devtools)
library(common)
# spatial data handling
library(terra)
library(sf)
# aesthetics
library(viridis)
library(webshot)
library(webshot2)
library(kableExtra)
library(patchwork)Note: I will be setting the global options of this
document so that only certain code chunks are rendered in the final
.html file. I will set the eval = FALSE so that none of the
code is re-run (preventing files from being overwritten during knitting)
and will simply overwrite this in chunks with plots.
I will load in preset styles for the maps. The regular map_style will be for the continuous suitability maps, while map_style_binarized will be used for specialized binary overlay maps between the regional ensemble and the global models.
map_style <- list(
xlab("UTM Easting"),
ylab("UTM Northing"),
theme_classic(),
theme(
# legend
legend.position = "bottom",
legend.title = element_text(margin = margin(r = 15, b = 45), hjust = 1),
legend.text = element_text(margin = margin(t = 10), angle = 35),
# background
panel.background = element_rect(fill = "lightblue2", colour = "lightblue2"),
# no axis titles
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank()
),
scale_x_continuous(expand = c(0, 0)),
scale_y_continuous(expand = c(0, 0)),
labs(fill = "Suitability for SLF \n(cloglog)"),
coord_equal()
)
map_style_binarized <- list(
xlab("UTM Easting"),
ylab("UTM Northing"),
theme_classic(),
theme(
# legend
legend.position = "bottom",
# background
panel.background = element_rect(fill = "lightblue2", colour = "lightblue2"),
# no axis titles
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank()
),
scale_x_continuous(expand = c(0, 0)),
scale_y_continuous(expand = c(0, 0)),
coord_equal()
)Import datasets
I will import the predicted suitability rasters and MTSS thresholds from each of the models. I will stack these rasters and harmonize their naming.
regional ensemble
# historical
regional_ensemble_1995 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_1981-2010.asc")
)
# CMIP6
# ssp126
regional_ensemble_2055_126 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp126.asc")
)
# ssp370
regional_ensemble_2055_370 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp370.asc")
)
# ssp585
regional_ensemble_2055_585 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp585.asc")
)
# the mean of the ssp scenarios
regional_ensemble_2055_ssp_mean <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp_averaged.asc")
)
regional_ensemble_thresholds <- read_csv(file = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_threshold_values.csv"))
# change names
names(regional_ensemble_1995) <- "ensemble_suitability"
# CMIP6
names(regional_ensemble_2055_126) <- "ensemble_suitability"
names(regional_ensemble_2055_370) <- "ensemble_suitability"
names(regional_ensemble_2055_585) <- "ensemble_suitability"
names(regional_ensemble_2055_ssp_mean) <- "ensemble_suitability"
# also convert to df
regional_ensemble_1995_df <- terra::as.data.frame(regional_ensemble_1995, xy = TRUE)
# CMIP6
regional_ensemble_2055_126_df <- terra::as.data.frame(regional_ensemble_2055_126, xy = TRUE)
regional_ensemble_2055_370_df <- terra::as.data.frame(regional_ensemble_2055_370, xy = TRUE)
regional_ensemble_2055_585_df <- terra::as.data.frame(regional_ensemble_2055_585, xy = TRUE)
regional_ensemble_2055_ssp_mean_df <- terra::as.data.frame(regional_ensemble_2055_ssp_mean, xy = TRUE)Global model
# historical
global_1995 <- terra::rast(
x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_1981-2010_mean.asc")
)
# CMIP6
# ssp126
global_2055_126 <- terra::rast(
x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_2041-2070_GFDL_ssp126_mean.asc")
)
# ssp370
global_2055_370 <- terra::rast(
x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_2041-2070_GFDL_ssp370_mean.asc")
)
# ssp126
global_2055_585 <- terra::rast(
x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_2041-2070_GFDL_ssp585_mean.asc")
)
# also import mean raster
global_2055_ssp_mean <- terra::rast(
x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_2041-2070_GFDL_ssp_averaged.asc")
)
summary_global <- read_csv(file.path(mypath, "slf_global_v4", "global_summary_all_iterations.csv")) %>%
dplyr::select(., statistic, mean)
# change names
# historical
names(global_1995) <- "global_suitability"
# CMIP6
names(global_2055_126) <- "global_suitability"
names(global_2055_370) <- "global_suitability"
names(global_2055_585) <- "global_suitability"
names(global_2055_ssp_mean) <- "global_suitability"
# also convert to df
global_1995_df <- terra::as.data.frame(global_1995, xy = TRUE)
# CMIP6
global_2055_126_df <- terra::as.data.frame(global_2055_126, xy = TRUE)
global_2055_370_df <- terra::as.data.frame(global_2055_370, xy = TRUE)
global_2055_585_df <- terra::as.data.frame(global_2055_585, xy = TRUE)
global_2055_ssp_mean_df <- terra::as.data.frame(global_2055_ssp_mean, xy = TRUE)1. Map cloglog suitability
Before I binarize these mapped predictions, I will plot raw suitability maps from each of the models.
1.1 Mapping helper function
I will try to reduce the amount of repeated code by using a mapping helper function containing the body of the plotting code. This part will remain relatively unchanged each time.
map_cloglog_output <- function(raster.df, raster.value.name, thresh.MTP, thresh.MTSS) {
# raster.df = the dataframe of the raster
# raster.value.name = the name of the raster values
# thresh.MTP = the df cell coordinates of the MTP thresh
# thresh.MTSS = the df cell coordinates of the MTSS thresh
ggplot() +
# default style object
map_style +
# plot raster
geom_raster(data = raster.df, aes(x = x, y = y, fill = raster.value.name)) +
# aesthetics
scale_fill_gradientn(
# the values of the color divisions: 0, MTP, slightly above MTP, MTSS, slightly above MTSS, 0.5, 0.75, 1
values = c(
0, # minimum
as.numeric(thresh.MTP), # MTP thresh
as.numeric(thresh.MTP) + 0.0000001, # some small amount more than the MTP
as.numeric(thresh.MTSS), # MTSS thresh
as.numeric(thresh.MTSS) + 0.0000001,
# the rest
0.5,
0.75,
1
),
# these colors taken from viridis color scale D
# the colors for the divisions in the order: 0, MTP, slightly above MTP, MTSS, slightly above MTSS, 0.5, 0.75, 1
colors = c("azure4", "azure4", "azure2", "azure2", "#481567FF", "#2D708EFF", "#3CBB75FF", "#FDE725FF"),
# the tick marks
breaks = c(0, as.numeric(thresh.MTP), as.numeric(thresh.MTSS), 0.5, 0.75, 1.00),
# labels for the tick marks
labels = c("unsuitable", "", paste0(round(as.numeric(thresh.MTSS), 2), "\n(MTSS thresh)"), 0.5, 0.75, 1),
limits = c(0, 1.0),
guide = guide_colorbar(frame.colour = "black", ticks.colour = "black", barwidth = 20, draw.ulim = TRUE, draw.llim = TRUE)
)
}1.2 Map cloglog output of models- historical
First, I will map the ensemble of the three regional-scale cloglog outputs.
regional_ensemble_1995_plot <- map_cloglog_output(
raster.df = regional_ensemble_1995_df, # the dataframe of the raster
raster.value.name = regional_ensemble_1995_df$ensemble_suitability, # the name of the raster values
thresh.MTP = regional_ensemble_thresholds[1, 4], # the df cell coordinates of the MTP thresh
thresh.MTSS = regional_ensemble_thresholds[6, 4] # the df cell coordinates of the MTSS thresh
) +
# labs
labs(
title = "Suitability for SLF in regional model ensemble",
subtitle = "1981-2010",
caption = "MTP & MTSS thresholds | clamped"
)
regional_ensemble_1995_plot## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.
ggsave(
regional_ensemble_1995_plot,
filename = file.path(mypath, "slf_regional_ensemble_v2", "plots", "regional_ensemble_pred_suit_globe_1981-2010.jpg"),
height = 8,
width = 12,
device = jpeg,
dpi = "retina"
)## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.
I will repeat the same process to map the predictions for each ssp scenario for 2041-2070. I will also mean these scenarios for a final prediction under climate change.
1.3 Map cloglog output of models- CMIP6
ssp mean
Finally, I will plot the mean of the three ssp scenarios. First, the regional ensemble.
## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.
Now I will plot the global-scale model.
## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.
2. Create binary threshold maps to infer range filling
Now that I have an idea of what the suitability maps look like before conversion, I will reclassify and combine these maps. I will create matrix objects that will be used to reclassify the output rasters to a binary predictor of suitability. I will use the MTSS (“Maximum Test Sensitivity Plus Specificity”) threshold for each model as the threshold for suitability, with anything below this threshold being considered unsuitable for SLF establishment.
To reclassify, package terra takes a 3-column matrix for its
classify() function, which re-classifies groups of values
to a single value. I will reclassify anything below the MTSS threshold
as one value, and anything above as another.
summary_global## # A tibble: 52 × 2
## statistic mean
## <chr> <dbl>
## 1 X.Training.samples 923.
## 2 Regularized.training.gain 3.78
## 3 Unregularized.training.gain 4.34
## 4 Iterations 2832
## 5 Training.AUC 0.988
## 6 X.Background.points 20000
## 7 bio11.contribution 52.2
## 8 bio12.contribution 19.0
## 9 bio15.contribution 21.7
## 10 bio2.contribution 7.08
## # ℹ 42 more rows
# MTSS value is 0.041
regional_ensemble_thresholds## # A tibble: 10 × 4
## thresh time_period CMIP6_model value
## <chr> <chr> <chr> <dbl>
## 1 MTP 1981-2010 NA 0.0114
## 2 MTP 2041-2070 ssp126_GFDL-ESM4 0.0114
## 3 MTP 2041-2070 ssp370_GFDL-ESM4 0.0115
## 4 MTP 2041-2070 ssp585_GFDL-ESM4 0.0115
## 5 MTP 2041-2070 ssp_mean_GFDL-ESM4 0.0115
## 6 MTSS 1981-2010 NA 0.180
## 7 MTSS 2041-2070 ssp126_GFDL-ESM4 0.179
## 8 MTSS 2041-2070 ssp370_GFDL-ESM4 0.179
## 9 MTSS 2041-2070 ssp585_GFDL-ESM4 0.179
## 10 MTSS 2041-2070 ssp_mean_GFDL-ESM4 0.179
# MTSS value is ~0.179I will create separate classification matrices for each of the global
and regional-scale ensemble outputs. The reason for having two separate
matrices is because I will need to differentiate between a “suitable”
spot on the global and regional rasters. Base-2 binary offers an
intuitive system for creating these class differences. I will encode the
rasters into base-2 binary so that the end result is 4 categories of
values (the idea for this was found in this
forum).
I will encode the cloglog output of MaxEnt into these values:
| Model and suitability | cloglog range | binary value | output value |
|---|---|---|---|
| Regional unsuitable | 0 - 0.241 | 00000001 | 1 |
| Regional suitable | 0.2411 - 1 | 00000010 | 2 |
| Global unsuitable | 0 - 0.041 | 00000100 | 4 |
| Global suitable | 0.0411 - 1 | 00001000 | 8 |
By encoding the rasters into base-2, I have created only 4 possible values of the raster layers, which can be translated into 4 categories:
- Unsuitable Agreement = 5 (00000101)
- Suitable Agreement = 10 (00001010)
- unsuitable regional/suitable global = 9 (00001001)
- Suitable regional/unsuitable global = 6 (00000110)
I will also interpret these categories in terms of risk of SLF suitability. These categories can be thought of as:
- Unsuitable Agreement = low risk
- Suitable Agreement = high risk
- Suitable regional/unsuitable global = adaptive risk
- unsuitable regional/suitable global = imminent risk
Gallien et al, 2012 interprets these categories in terms of specific ecological mechanisms, but we prefer to interpret them in terms of the risk for SLF.
2.1 convert output rasters to binary predictors
I will walk through this workflow initially, but will functionalize it after this to avoid repeating it four more times.
historical
To convert the rasters to binary predictors of presence / absence,
first I will create the classification matrices needed by the
classify() function. Before converting, lets check the
range of values.
# get the range of the values in the raster
terra::minmax(global_1995) %>%
format(., scientific = FALSE) # convert scientific notation to decimals## global_suitability
## min "0.0000000003152319"
## max "1.0000000000000000"
# get the range of the values in the raster
terra::minmax(regional_ensemble_1995) %>%
format(., scientific = FALSE)## ensemble_suitability
## min "0.0000001860083"
## max "0.7682798504829"
# all looks goodNow I will create the classification rasters. I will pull these from their respective .csv files.
# create regional suitability value matrix for terra
regional_ensemble_classes <- data.frame(
from = c(0, as.numeric(regional_ensemble_thresholds[6, 4]) + 0.0000000001), # add a small amount to the lower end of the 2nd range
to = c(as.numeric(regional_ensemble_thresholds[6, 4]), 1),
becomes = c(strtoi(00000001, base = 2), strtoi(00000010, base = 2)) # the strtoi function converts to base-2, becomes 1 and 2
)
global_classes <- data.frame(
from = c(0, as.numeric(summary_global[42, 2]) + 0.0000000001),
to = c(as.numeric(summary_global[42, 2]), 1),
becomes = c(strtoi(00000100, base = 2), strtoi(00001000, base = 2)) # becomes 4 and 8
)
regional_ensemble_classes## from to becomes
## 1 0.0000000 0.1795021 1
## 2 0.1795021 1.0000000 2
global_classes## from to becomes
## 1 0.00000 0.01693 4
## 2 0.01693 1.00000 8Now I can reclassify the MaxEnt outputs. I will load in the averaged predictions for the global and regional models for the time period 1981-2010. I will reclassify their values based on the two matrices above. This chunk might take some time to run.
# regional
# reclassify and write regional raster
regional_ensemble_1995_classified <- terra::classify(
x = regional_ensemble_1995,
rcl = regional_ensemble_classes,
right = NA, # close both ends of the range of classification values
others = strtoi(00000001, base = 2), # make it the unsuitable value if not in this range
filename = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_1981-2010.asc"), # also write to file
filetype = "AAIGrid",
overwrite = FALSE
)
# global
# reclassify and write global raster
global_1995_classified <- terra::classify(
x = global_1995,
rcl = global_classes,
right = NA,
others = strtoi(00000100, base = 2),
filename = file.path(mypath, "working_dir", "slf_global_v4_binarized_1981-2010.asc"),
filetype = "AAIGrid",
overwrite = FALSE
) I will run a few error checks to ensure that the binarization process worked.
# take minmax
terra::minmax(global_1995_classified) %>%
format(., scientific = FALSE)
terra::minmax(regional_ensemble_1995_classified) %>%
format(., scientific = FALSE)
# all good
# check unique values
unique(values(global_1995_classified))
unique(values(regional_ensemble_1995_classified))Now that the rasters have been converted to binary, we will visualize them to ensure that the process worked. Since the same process was done to both the regional and global models, I will only plot the regional model.
# load in binary raster just created and convert to df
regional_ensemble_1995_classified_df <- terra::rast(
x = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_1981-2010.asc")
) %>%
terra::as.data.frame(., xy = TRUE) #%>%
#na.omit()
# now, create vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- unique(regional_ensemble_1995_classified_df$ensemble_suitability)
# labels for the values
labels.obj <- c("regional unsuitable", "regional suitable")
# vector of colors to classify scale
values.obj <- c(
"azure4",
"darkblue"
)
# plot regional binary
ggplot() +
geom_raster(data = regional_ensemble_1995_classified_df, aes(x = x, y = y, fill = as.factor(ensemble_suitability))) +
labs(
title = "Binarized suitability for 'regional_ensemble' model",
subtitle = "1981-2010"
) +
scale_discrete_manual(
name = "suitability for SLF",
values = values.obj,
breaks = breaks.obj,
labels = labels.obj,
aesthetics = "fill",
) +
map_style_binarized## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.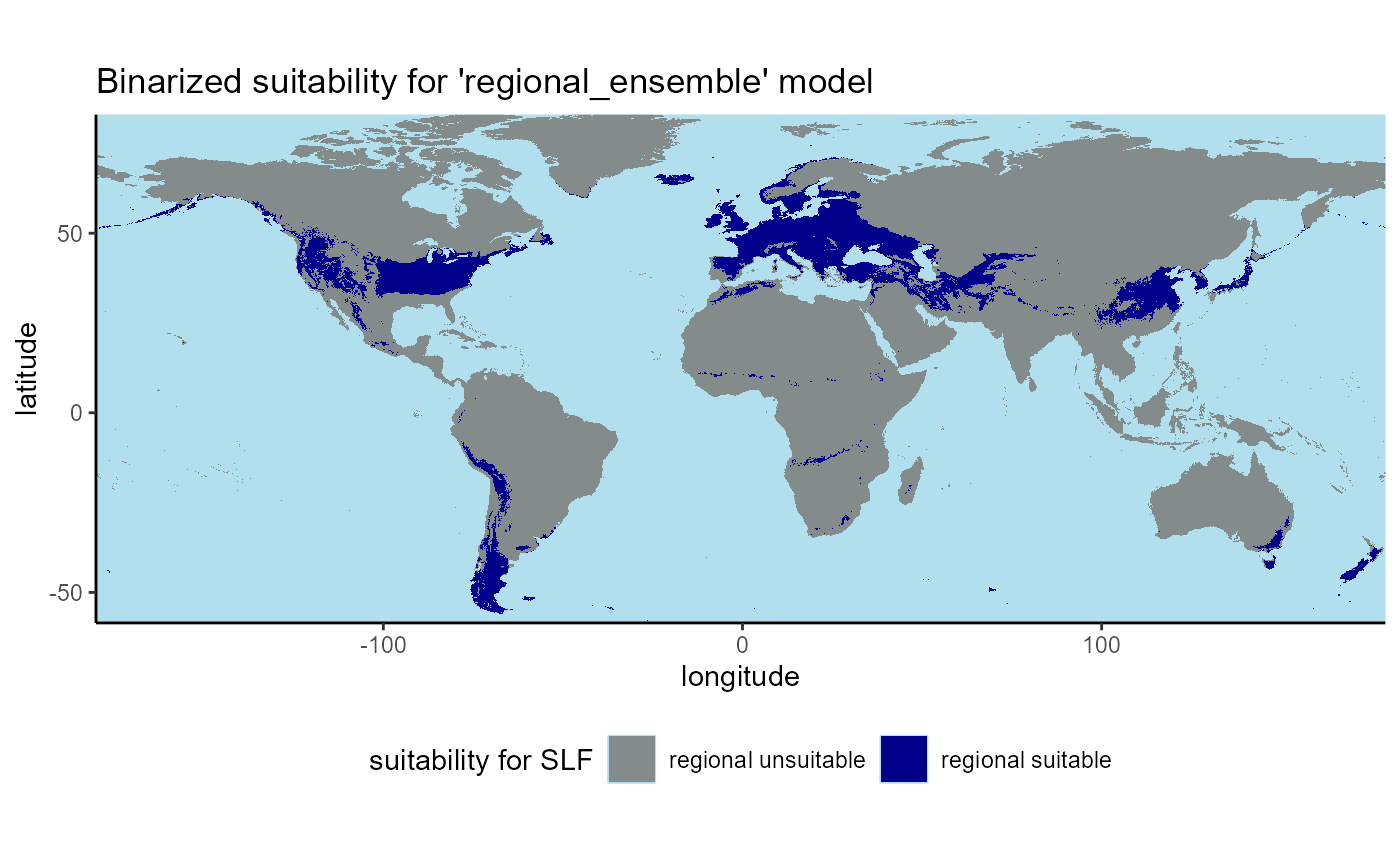
I will perform the same operation as above for the predicted climate change rasters from 2041-2070 (ssp126, 370, 585 and the mean of these).
The helper function below, classify_matrix(), will
create the object needed to classify each raster into the binary-based
classes I outlined above.
helper function- terra classify
classify_matrix <- function(thresh, model.scale) {
# thresh = the threshold for suitability in that model
# model = either of "global" or "regional"
# the strtoi function converts a number to base-2
# regional model classes
if (model.scale == "regional") {
data.frame(
from = c(0, as.numeric(thresh) + 0.0000000001), # add a small amount to the lower end of the 2nd range
to = c(as.numeric(thresh), 1),
becomes = c(strtoi(00000001, base = 2), strtoi(00000010, base = 2)) # becomes 1 and 2
)
# otherwise, go with global model
} else if (model.scale == "global") {
data.frame(
from = c(0, as.numeric(thresh) + 0.0000000001),
to = c(as.numeric(thresh), 1),
becomes = c(strtoi(00000100, base = 2), strtoi(00001000, base = 2)) # becomes 4 and 8
)
}
}ssp mean
# get the range of the values in the raster
terra::minmax(global_2055_ssp_mean) %>%
format(., scientific = FALSE) # convert scientific notation to decimals## global_suitability
## min "0.0000000006902004"
## max "1.0000000000000000"
# get the range of the values in the raster
terra::minmax(regional_ensemble_2055_ssp_mean) %>%
format(., scientific = FALSE)## ensemble_suitability
## min "0.0000001855588"
## max "0.7522506117821"
# global
global_classes <- classify_matrix(
thresh = summary_global[42, 2],
model.scale = "global"
)
# regional ensemble
regional_ensemble_classes <- classify_matrix(
thresh = regional_ensemble_thresholds[10, 4],
model.scale = "regional"
)
# regional
# reclassify and write regional raster
regional_ensemble_2055_ssp_mean_classified <- terra::classify(
x = regional_ensemble_2055_ssp_mean,
rcl = regional_ensemble_classes,
right = NA, # close both ends of the range of classification values
others = strtoi(00000001, base = 2), # make it the unsuitable value if not in this range
filename = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_2041-2070_ssp_mean_GFDL.asc"), # also write to file
filetype = "AAIGrid",
overwrite = FALSE
)
# global
# reclassify and write global raster
global_2055_ssp_mean_classified <- terra::classify(
x = global_2055_ssp_mean,
rcl = global_classes,
right = NA,
others = strtoi(00000100, base = 2),
filename = file.path(mypath, "working_dir", "slf_global_v4_binarized_2041-2070_ssp_mean_GFDL.asc"),
filetype = "AAIGrid",
overwrite = FALSE
)
# take minmax
terra::minmax(global_2055_ssp_mean_classified) %>%
format(., scientific = FALSE)
terra::minmax(regional_ensemble_2055_ssp_mean_classified) %>%
format(., scientific = FALSE)
# all good
# check unique values
unique(values(global_2055_ssp_mean_classified))
unique(values(regional_ensemble_2055_ssp_mean_classified))Now lets plot the binarized mean of the ssp scenarios.
# load in binary raster just created and convert to df
regional_ensemble_2055_ssp_mean_classified_df <- terra::rast(
x = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_2041-2070_ssp_mean_GFDL.asc")
) %>%
terra::as.data.frame(., xy = TRUE)
# now, create vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- unique(regional_ensemble_2055_ssp_mean_classified_df$ensemble_suitability)
# labels for the values
labels.obj <- c("regional unsuitable", "regional suitable")
# vector of colors to classify scale
values.obj <- c(
"azure4",
"darkblue"
)
# plot regional binary
ggplot() +
geom_raster(data = regional_ensemble_2055_ssp_mean_classified_df, aes(x = x, y = y, fill = as.factor(ensemble_suitability))) +
labs(
title = "Binarized suitability for 'regional_ensemble' model",
subtitle = "projected to 2041-2070 | mean of ssp126, 370, 585 | GFDL-ESM4"
) +
scale_discrete_manual(
name = "suitability for SLF",
values = values.obj,
breaks = breaks.obj,
labels = labels.obj,
aesthetics = "fill"
) +
map_style_binarized## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.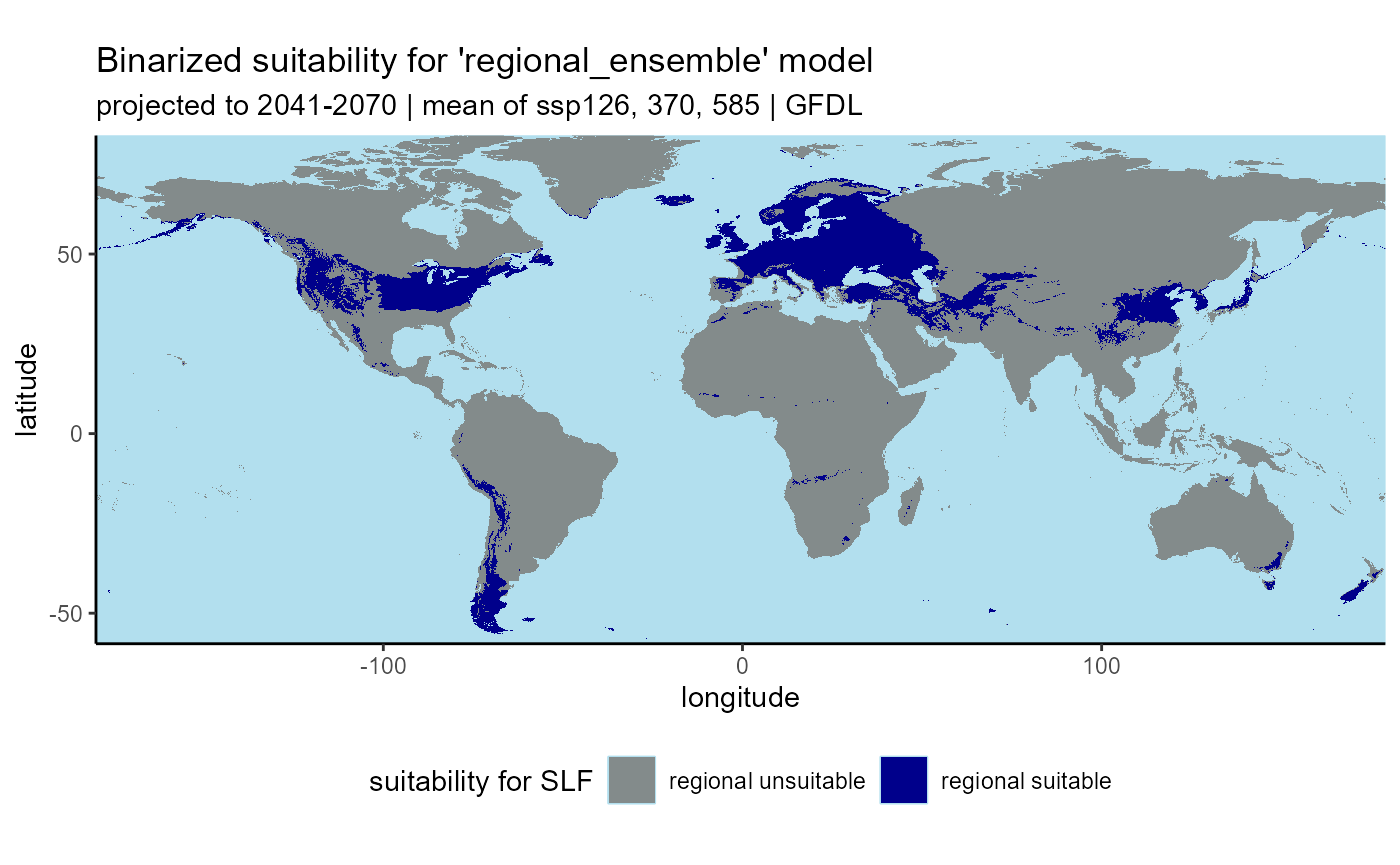
2.2 Mosaic rasters and produce map
From the plots, we see that the binary conversion worked. Next, I will need to stack the reclassified global and regional rasters and add them into a single raster output. This should give me a final raster with 4 classes:
| SLF suitability | raster value |
|---|---|
| Unsuitable Agreement | 5 |
| Suitable Agreement | 10 |
| unsuitable regional/suitable global | 9 |
| Suitable regional/unsuitable global | 6 |
The mosaic function combines spatRasters that overlap in extent into a new raster, via the function specified (addition, in this case). I will run through this workflow for the historical rasters, then will functionalize the plotting for the ssp scenarios.
historical
global_1995_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_global_v4_binarized_1981-2010.asc"))
regional_ensemble_1995_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_1981-2010.asc"))
# overlay and combine rasters by summing values
slf_binarized_1995 <- terra::mosaic(
global_1995_classified, regional_ensemble_1995_classified, # rasters
fun = "sum",
filename = file.path(mypath, "working_dir", "slf_binarized_summed_1981-2010.asc"),
overwrite = FALSE,
wopt = list(progress = 1)) # progress barI will prepare plotting objects.
slf_binarized_1995 <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_1981-2010.asc"))
# rename values
names(slf_binarized_1995) <- "global_regional_binarized"
# convert to df
slf_binarized_1995_df <- terra::as.data.frame(slf_binarized_1995, xy = TRUE) #%>%
# na.omit()
# check for the number of unique values in the raster values column
unique(slf_binarized_1995_df$global_regional_binarized)## [1] 5 6 10 9
# it seems that the reclassification worked! Lets map it to be sure.
# now, create vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- c(5, 9, 6, 10) # i manually ordered these
# labels for the values
labels.obj <- c("low", "moderate", "high", "extreme")
# vector of colors to classify scale
values.obj <- c("azure4", "gold", "darkorange", "darkred")
slf_binarized_1995_plot <- ggplot() +
geom_raster(data = slf_binarized_1995_df, aes(x = x, y = y, fill = as.factor(global_regional_binarized))) +
labs(
title = "Projected risk of Lycorma delicatula invasion globally",
subtitle = "1981-2010"
) +
scale_discrete_manual(
name = "projected risk",
values = values.obj,
breaks = breaks.obj,
labels = labels.obj,
aesthetics = "fill",
guide = guide_colorsteps(frame.colour = "black", ticks.colour = "black", barwidth = 20, draw.ulim = TRUE, draw.llim = TRUE)
) +
map_style_binarized +
guides(fill = guide_legend(nrow = 1, byrow = TRUE)) +
theme(legend.key = element_rect(color = "black"))
slf_binarized_1995_plot## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.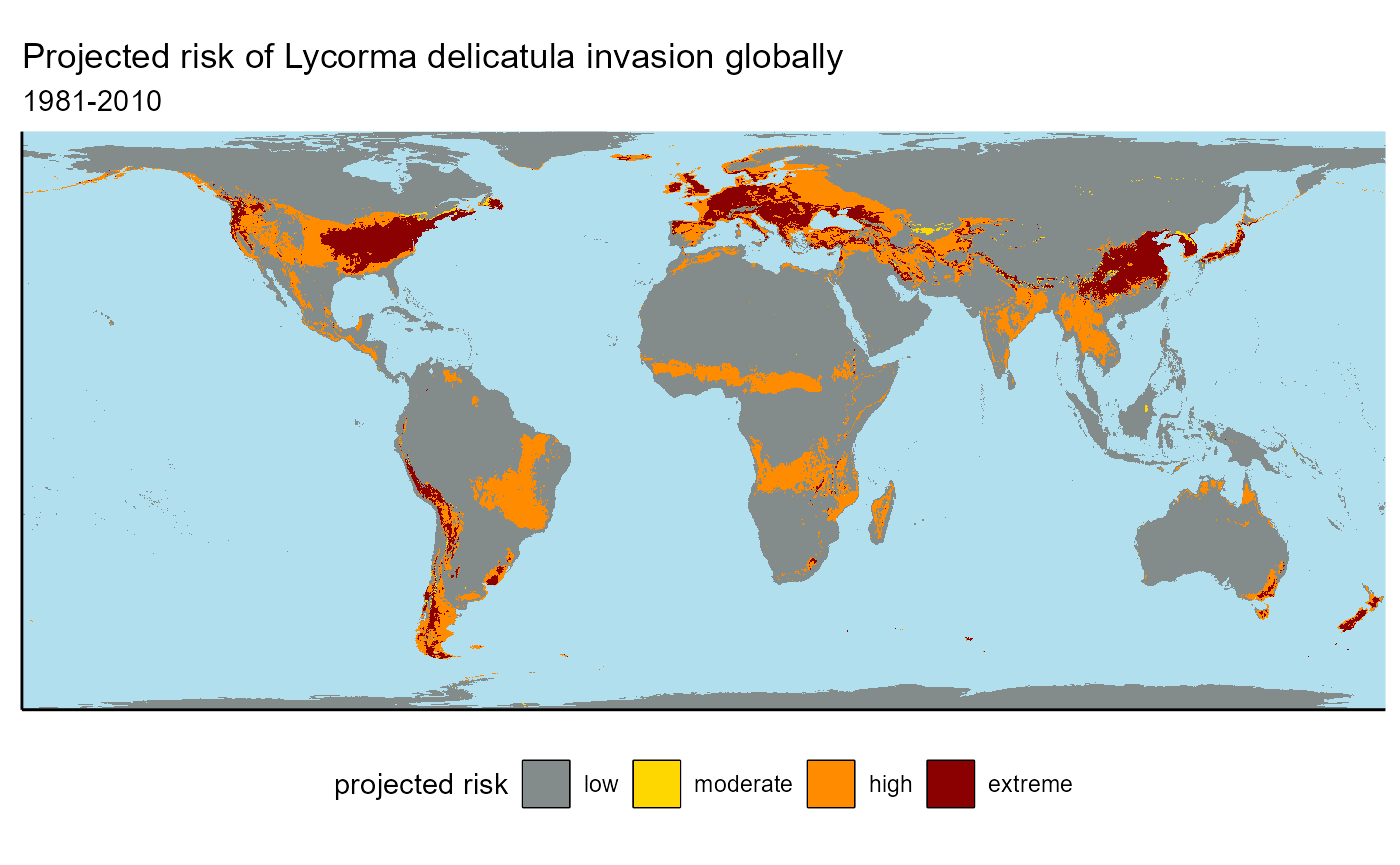
helper function- binary raster plotting
Now that I have run through the process once, I will functionalize the plotting process for the ssp scenarios to save time and lines of code.
# first, create vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- c(5, 9, 6, 10) # i manually ordered these
# labels for the values
labels.obj <- c("low", "moderate", "high", "extreme")
# vector of colors to classify scale
values.obj <- c("azure4", "gold", "darkorange", "darkred")
# function
plot_binary <- function(raster.df) {
# raster.df = data frame of the raster to be plotted
ggplot() +
# default map style
map_style_binarized +
# raster layer
geom_raster(data = raster.df, aes(x = x, y = y, fill = as.factor(global_regional_binarized))) +
# custom discrete scale
scale_discrete_manual(
name = "projected risk",
# use objects outlined above
values = values.obj,
breaks = breaks.obj,
labels = labels.obj,
aesthetics = "fill",
guide = guide_colorsteps(frame.colour = "black", ticks.colour = "black", barwidth = 20, draw.ulim = TRUE, draw.llim = TRUE)
) +
# other theme customizations
guides(fill = guide_legend(nrow = 1, byrow = TRUE)) +
theme(legend.key = element_rect(color = "black"))
}ssp mean
global_2055_ssp_mean_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_global_v4_binarized_2041-2070_ssp_mean_GFDL.asc"))
regional_ensemble_2055_ssp_mean_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_regional_ensemble_v2_binarized_2041-2070_ssp_mean_GFDL.asc"))
# overlay and combine rasters by summing values
slf_binarized_2055_ssp_mean <- terra::mosaic(
global_2055_ssp_mean_classified, regional_ensemble_2055_ssp_mean_classified, # rasters
fun = "sum",
filename = file.path(mypath, "working_dir", "slf_binarized_summed_2041-2070_ssp_mean_GFDL.asc"),
overwrite = FALSE,
wopt = list(
progress = 1, # progress bar
names = "global_regional_binarized" # layer name, change in function if this changes
)
)
slf_binarized_2055_ssp_mean <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_2041-2070_ssp_mean_GFDL.asc"))
# convert to df
slf_binarized_2055_ssp_mean_df <- terra::as.data.frame(slf_binarized_2055_ssp_mean, xy = TRUE) #%>%
# na.omit()
# check for the number of unique values in the raster values column
unique(slf_binarized_2055_ssp_mean_df$global_regional_binarized)## [1] 5 6 10 9
# it seems that the reclassification worked! Lets map it to be sure.
slf_binarized_2055_ssp_mean_plot <- plot_binary(
raster.df = slf_binarized_2055_ssp_mean_df
) +
# labels
labs(
title = "Projected risk of Lycorma delicatula invasion globally under climate change",
subtitle = "2041-2070 | mean of ssp126, ssp370 and ssp585 | GFDL-ESM4"
)
slf_binarized_2055_ssp_mean_plot## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.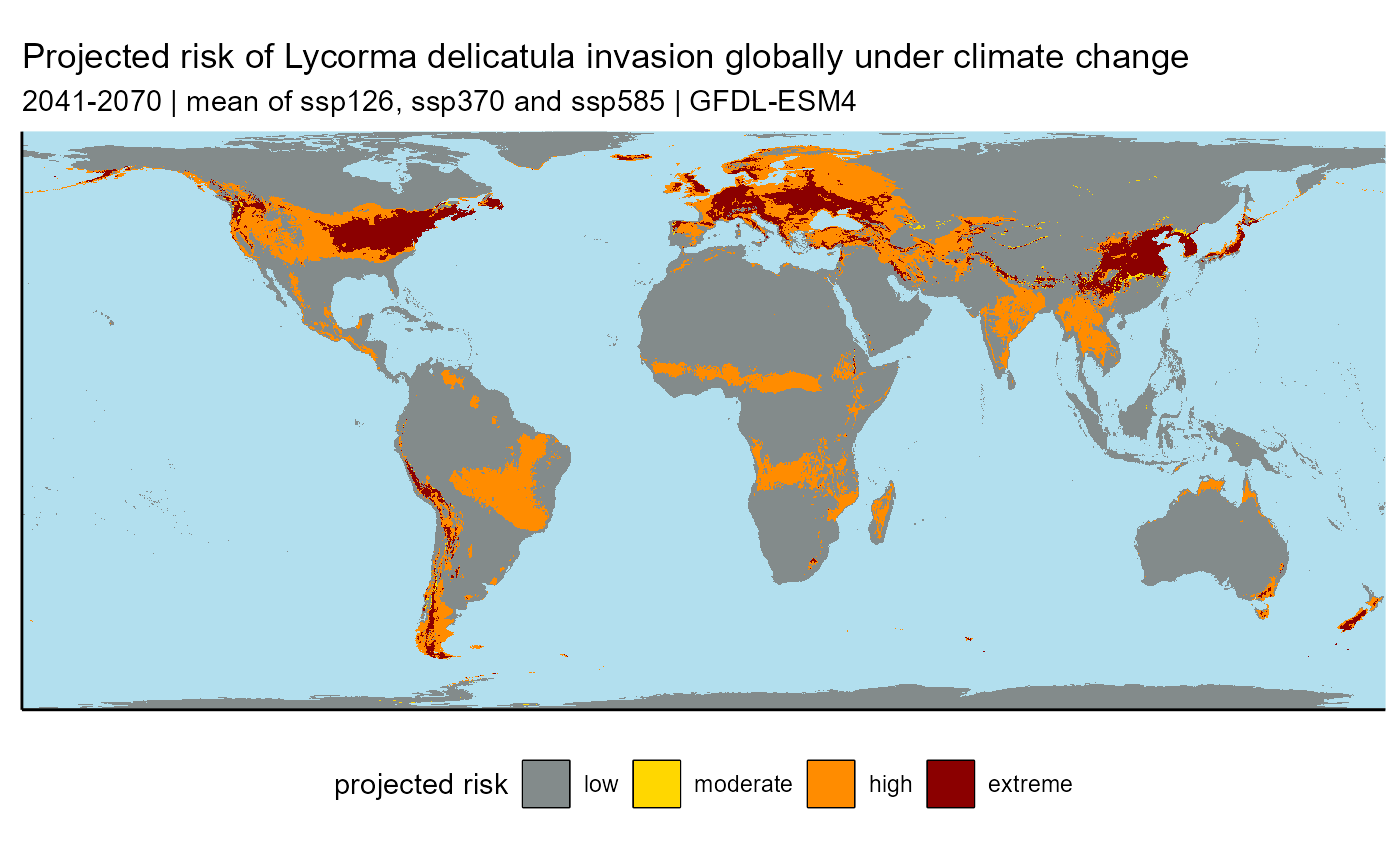
3. Calculate total area predicted by each model
The figures above are classified into categories of low, moderate, high and extreme risk for SLF establishment. In addition to visualizing them, I want to know the total predicted area in each category.
I will calculate the total suitable area predicted by each category and model.
As a reminder, the categories in these rasters are:
| SLF suitability | raster value |
|---|---|
| Unsuitable Agreement | 5 |
| Suitable Agreement | 10 |
| unsuitable regional/suitable global | 9 |
| Suitable regional/unsuitable global | 6 |
# load rasters
slf_binarized_1995 <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_1981-2010.asc"))
slf_binarized_2055_ssp_mean <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_2041-2070_ssp_mean_GFDL.asc"))
# 1995
slf_model_prop_table_1995 <- terra::expanse(
x = slf_binarized_1995,
unit = "km",
byValue = TRUE # group by cell value, which is the risk level
)
# 2055
slf_model_prop_table_2055_ssp_mean <- terra::expanse(
x = slf_binarized_2055_ssp_mean,
unit = "km",
byValue = TRUE
)
# NOTE this throws GDAL error 2050 "reprojection failed"- I read this means that some points are outside of projection domain- this happened initially when I projected the original rasters to ESRI:54017, so I am guessing this is related to that issue
# create list of range shift values
categories.obj <- tibble(
model_suitability = c("unsuitable_agreement", "regional", "global", "suitable_agreement"),
value = c(5, 6, 9, 10)
)
# join
slf_model_prop_table_joined <- left_join(slf_model_prop_table_1995, slf_model_prop_table_2055_ssp_mean, by = c("value", "layer")) %>%
# add labels
left_join(., categories.obj, by = "value")
# tidy
slf_model_prop_table_joined <- slf_model_prop_table_joined %>%
dplyr::select(-c(layer, value)) %>%
dplyr::rename(
"area_km_1995" = "area.x",
"area_km_2055" = "area.y"
) %>%
dplyr::mutate(
# make proportions
prop_total_area_1995 = scales::label_percent()(area_km_1995 / sum(area_km_1995)),
prop_total_area_2055 = scales::label_percent()(area_km_2055 / sum(area_km_2055)),
# change labels
area_km_1995 = scales::label_comma()(area_km_1995),
area_km_2055 = scales::label_comma()(area_km_2055)
) %>%
dplyr::relocate(model_suitability, area_km_1995, prop_total_area_1995, area_km_2055, prop_total_area_2055) %>%
dplyr::rename(
"prop_area_present" = "prop_total_area_1995",
"prop_area_2041-2070" = "prop_total_area_2055"
)
# add superscript
colnames(slf_model_prop_table_joined)[2] <- paste0("area_km", common::supsc("2"), "_present")
colnames(slf_model_prop_table_joined)[4] <- paste0("area_km", common::supsc("2"), "_2041-2070")
# .html formatting
# format row colors
slf_model_prop_table_joined[1, 1] <- kableExtra::cell_spec(slf_model_prop_table_joined[1, 1], format = "html", bold = TRUE, color = "azure4")
slf_model_prop_table_joined[2, 1] <- kableExtra::cell_spec(slf_model_prop_table_joined[2, 1], format = "html", bold = TRUE, color = "darkorange")
slf_model_prop_table_joined[3, 1] <- kableExtra::cell_spec(slf_model_prop_table_joined[3, 1], format = "html", bold = TRUE, color = "gold")
slf_model_prop_table_joined[4, 1] <- kableExtra::cell_spec(slf_model_prop_table_joined[4, 1], format = "html", bold = TRUE, color = "darkred")
# convert to kable
slf_model_prop_kable <- knitr::kable(x = slf_model_prop_table_joined, format = "html", escape = FALSE) %>%
kableExtra::kable_styling(bootstrap_options = "striped", full_width = TRUE)
print(slf_model_prop_kable)## <table class="table table-striped" style="margin-left: auto; margin-right: auto;">
## <thead>
## <tr>
## <th style="text-align:left;"> model_suitability </th>
## <th style="text-align:left;"> area_km²_present </th>
## <th style="text-align:left;"> prop_area_present </th>
## <th style="text-align:left;"> area_km²_2041-2070 </th>
## <th style="text-align:left;"> prop_area_2041-2070 </th>
## </tr>
## </thead>
## <tbody>
## <tr>
## <td style="text-align:left;"> <span style=" font-weight: bold; color: rgba(131, 139, 139, 255) !important;">unsuitable_agreement</span> </td>
## <td style="text-align:left;"> 120,739,040 </td>
## <td style="text-align:left;"> 79.3% </td>
## <td style="text-align:left;"> 118,753,204 </td>
## <td style="text-align:left;"> 78.0% </td>
## </tr>
## <tr>
## <td style="text-align:left;"> <span style=" font-weight: bold; color: darkorange !important;">regional</span> </td>
## <td style="text-align:left;"> 21,290,127 </td>
## <td style="text-align:left;"> 14.0% </td>
## <td style="text-align:left;"> 23,792,459 </td>
## <td style="text-align:left;"> 15.6% </td>
## </tr>
## <tr>
## <td style="text-align:left;"> <span style=" font-weight: bold; color: gold !important;">global</span> </td>
## <td style="text-align:left;"> 521,482 </td>
## <td style="text-align:left;"> 0.3% </td>
## <td style="text-align:left;"> 446,947 </td>
## <td style="text-align:left;"> 0.3% </td>
## </tr>
## <tr>
## <td style="text-align:left;"> <span style=" font-weight: bold; color: darkred !important;">suitable_agreement</span> </td>
## <td style="text-align:left;"> 9,759,184 </td>
## <td style="text-align:left;"> 6.4% </td>
## <td style="text-align:left;"> 9,317,225 </td>
## <td style="text-align:left;"> 6.1% </td>
## </tr>
## </tbody>
## </table>
# model training dataset
# save as .csv
write_csv(slf_model_prop_table_joined, file.path(here::here(), "vignette-outputs", "data-tables", "slf_binarized_summed_areas_table.csv"))
# save as .html
kableExtra::save_kable(
slf_model_prop_kable,
file = file.path(here::here(), "vignette-outputs", "figures", "slf_binarized_summed_areas_table.html"),
self_contained = TRUE,
bs_theme = "simplex"
)
# initialize webshot by
# webshot::install_phantomjs()
# convert to jpg
webshot::webshot(
url = file.path(here::here(), "vignette-outputs", "figures", "slf_binarized_summed_areas_table.html"),
file = file.path(here::here(), "vignette-outputs", "figures", "slf_binarized_summed_areas_table.jpg"),
zoom = 2
)
# remove html
file.remove(file.path(here::here(), "vignette-outputs", "figures", "slf_binarized_summed_areas_table.html"))4. Map range shifting areas
Lastly, I want to create a map of areas where SLF range is expanding, contracting or remaining the same over time. I will binarize the rasters so that any point that is above the MTSS threshold on either model becomes one number and anything above the threshold becomes another. This will follow a slightly altered version of the methodology from step 2.
I will classify the cells as the following:
| Raster and suitability | cloglog range | binary value | output value |
|---|---|---|---|
| Historical unsuitable | 5 | 00000001 | 1 |
| Historical suitable | 6, 9 or 10 | 00000010 | 2 |
| 2041-2070 unsuitable | 5 | 00000100 | 4 |
| 2041-2070 suitable | 6, 9 or 10 | 00001000 | 8 |
By encoding the rasters into base-2, I have created only 4 possible values of the raster layers, which can be translated into 4 categories. I will also interpret these categories in terms of risk of SLF range expansion::
| Suitability agreement | summed value | binary value | interpretation |
|---|---|---|---|
| Unsuitable Agreement | 5 | 00000101 | will stay unsuitable (no range shift) |
| Suitable Agreement | 10 | 00001010 | will stay suitable (no range shift) |
| unsuitable hist/suitable 2041-2070 | 9 | 00001001 | will become SUITABLE in the future (range expansion) |
| Suitable hist/unsuitable 2041-2070 | 6 | 00000110 | will become UNSUITABLE in the future (range contraction) |
I will classify areas of unsuitability as gray, per usual. Areas where SLF remains the same will be white, areas where SLF is expanding will be green, and areas where SLF is contracting will be red.
First, I will load in the historical binarized raster (the intersection between the regional and global models) and classify it in preparation for this analysis. Then, I will intersect this raster with each of the predicted ssp rasters and the mean of those scenarios.
4.1 Setup
# 1995
slf_binarized_1995 <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_1981-2010.asc"))
# rename values
names(slf_binarized_1995) <- "global_regional_binarized_1995"
# convert to df
slf_binarized_1995_df <- terra::as.data.frame(slf_binarized_1995, xy = TRUE) I will also load some classification matrices for
terra::classify(). These should not change between maps
because we are classifying based on the time period, which does not
change.
# create historical suitability value matrix for terra
classes_1995 <- data.frame(
is = c(5, 6, 9, 10),
becomes = c(strtoi(00000001, base = 2), strtoi(00000010, base = 2), strtoi(00000010, base = 2), strtoi(00000010, base = 2))
# the strtoi function converts to base-2, becomes 1 and 2
)
# the class for all CMIP6 scenarios
classes_2055 <- data.frame(
is = c(5, 6, 9, 10),
becomes = c(strtoi(00000100, base = 2), strtoi(00001000, base = 2), strtoi(00001000, base = 2), strtoi(00001000, base = 2))
# becomes 4 and 8
)
classes_1995## is becomes
## 1 5 1
## 2 6 2
## 3 9 2
## 4 10 2
classes_2055## is becomes
## 1 5 4
## 2 6 8
## 3 9 8
## 4 10 8I will go ahead and classify the historical binarized raster because I will use it in the next steps
# reclassify and write the historical binarized raster
slf_binarized_1995_classified <- terra::classify(
x = slf_binarized_1995,
rcl = classes_1995,
others = strtoi(00000001, base = 2),
filename = file.path(mypath, "working_dir", "slf_range_shift_1981-2010.asc"), # also write to file
filetype = "AAIGrid",
overwrite = FALSE
) I will quickly ensure that the binarization process worked by creating a rough plot.
# load in binary raster just created and convert to df
slf_binarized_1995_classified_df <- terra::rast(
x = file.path(mypath, "working_dir", "slf_range_shift_1981-2010.asc")
) %>%
terra::as.data.frame(., xy = TRUE) #%>%
#na.omit()
# now, create vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- unique(slf_binarized_1995_classified_df$global_regional_binarized_1995)
# labels for the values
labels.obj <- c("present unsuitable", "present suitable")
# vector of colors to classify scale
values.obj <- c(
"azure4",
"azure"
)
# plot regional binary
ggplot() +
geom_raster(data = slf_binarized_1995_classified_df, aes(x = x, y = y, fill = as.factor(global_regional_binarized_1995))) +
labs(title = "current suitability for SLF") +
scale_fill_manual(
name = "suitability for SLF",
values = values.obj,
breaks = breaks.obj,
labels = labels.obj,
aesthetics = "fill",
) +
map_style_binarizedThe conversion worked, as anything colorful on the binarized map should now be white. I will load a helper function for future plotting, similar to the one in step 2.2. I will interpret these categories in terms range shifts, as discussed above:
- Unsuitable Agreement = 5 -> area will stay unsuitable (no range shift)
- Suitable Agreement = 10 -> area will stay suitable (no range shift)
- unsuitable 1995/suitable 2055 = 9 -> area will become SUITABLE in the future (range expansion)
- Suitable 1995/unsuitable 2055 = 6 -> area will become UNSUITABLE in the future (range contraction)
# vectors of values used to manually edit the scale of the plot
# the possible values of the scale and their order
breaks.obj <- c(5, 10, 6, 9) # i manually ordered these according to the order produced from unique(slf_range_shift_126_df$range_shift_summed)
# labels for the values
labels.obj <- c("unsuitable area\nretained", "suitabile area\nretained", "contraction\nsuitable area", "expansion\nsuitable area")
# vector of colors to classify scale
values.obj <- c("azure4", "azure", "darkblue", "darkgreen")
# function
plot_range_shift <- function(raster.df) {
# raster.df = the df of the raster to be plotted
ggplot() +
# default style
map_style_binarized +
# plot raster
geom_raster(data = raster.df, aes(x = x, y = y, fill = as.factor(range_shift_summed))) +
# discretized scale
scale_discrete_manual(
name = "L delicatula\nsuitability shift",
values = values.obj, # the colors
breaks = breaks.obj, # the breaks in the colors (4 since this is a discrete scale)
labels = labels.obj, # the labels per color bin
aesthetics = "fill",
guide = guide_colorsteps(frame.colour = "black", ticks.colour = "black", barwidth = 20, draw.ulim = TRUE, draw.llim = TRUE)
) +
# aesthetics
guides(fill = guide_legend(nrow = 1, byrow = TRUE)) +
theme(legend.key = element_rect(color = "black"), legend.title = element_text(hjust = 1))
}I will create another helper function to format a range shift table per ssp scenario, which I will create later.
# create list of range shift values
ranges.obj <- c("remains_unsuitable", "contraction", "expansion", "retained_suitability")
table_range_shift <- function(table.df) {
# table.df = dataframe of areas for raster bins, created by terra::expanse
# tidy
table.df.output <- table.df %>%
dplyr::select(-layer) %>%
dplyr::mutate(
Ld_range_shift_type = ranges.obj,
prop_total_area = scales::label_percent()(area / sum(area)),
area = scales::label_comma()(area)
) %>%
dplyr::select(-value) %>%
dplyr::relocate(Ld_range_shift_type, area)
colnames(table.df.output)[2] <- paste0("area_km", common::supsc("2"))
# .html formatting
# format row colors
table.df.output[1, 1] <- kableExtra::cell_spec(table.df.output[1, 1], format = "html", background = "azure4")
table.df.output[2, 1] <- kableExtra::cell_spec(table.df.output[2, 1], format = "html", background = "blue")
table.df.output[3, 1] <- kableExtra::cell_spec(table.df.output[3, 1], format = "html", background = "darkgreen")
table.df.output[4, 1] <- kableExtra::cell_spec(table.df.output[4, 1], format = "html", background = "azure")
# return
return(table.df.output)
}4.2 ssp 126
classify
slf_binarized_2055_126 <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_2041-2070_ssp126_GFDL.asc"))
# rename values
names(slf_binarized_2055_126) <- "global_regional_binarized_2055_126"
# convert to df
slf_binarized_2055_126_df <- terra::as.data.frame(slf_binarized_2055_126, xy = TRUE)
# global
# reclassify and write binarized raster
slf_binarized_2055_126_classified <- terra::classify(
x = slf_binarized_2055_126,
rcl = classes_2055,
others = strtoi(00000100, base = 2),
filename = file.path(mypath, "working_dir", "slf_range_shift_2041-2070_ssp126_GFDL.asc"),
filetype = "AAIGrid",
overwrite = FALSE
) I will run a few error checks to ensure that the binarization process worked.
# take minmax
terra::minmax(slf_binarized_1995_classified) %>%
format(., scientific = FALSE)
terra::minmax(slf_binarized_2055_126_classified) %>%
format(., scientific = FALSE)
# all good
# check unique values
unique(values(slf_binarized_1995_classified))
unique(values(slf_binarized_2055_126_classified))Mosaic
Next, I will need to stack the reclassified 1995 and 2055 rasters and add them into a single raster output. This should give me the a raster with 4 classes:
- Unsuitable Agreement = 5
- Suitable Agreement = 10
- unsuitable 1995/suitable 2055 = 9
- Suitable 1995/unsuitable 2055 = 6
The mosaic function combines spatRasters that overlap in extent into a new raster, via the function specified (addition, in this case).
slf_binarized_1995_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_1981-2010.asc"))
slf_binarized_2055_126_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_2041-2070_ssp126_GFDL.asc"))
# overlay and combine rasters by summing values
slf_range_shift_126 <- terra::mosaic(
slf_binarized_1995_classified, slf_binarized_2055_126_classified, # rasters
fun = "sum",
filename = file.path(mypath, "working_dir", "slf_range_shift_summed_ssp126_GFDL.asc"),
overwrite = FALSE,
wopt = list(
progress = 1,
names = "range_shift_summed"
)
) # progress barPlot
# re-load created raster
slf_range_shift_126 <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_summed_ssp126_GFDL.asc"))
# convert to df
slf_range_shift_126_df <- terra::as.data.frame(slf_range_shift_126, xy = TRUE) #%>%
# na.omit()
# check for the number of unique values in the raster values column
unique(slf_range_shift_126_df$range_shift_summed)## [1] 5 9 10 6
# it seems that the reclassification worked! Lets map it to be sure.
slf_range_shift_126_plot <- plot_range_shift(
raster.df = slf_range_shift_126_df
) +
# labels
labs(
title = "Projected areas suitable for Lycorma delicatula range expansion",
subtitle = "2041-2070 | ssp126 | GFDL-ESM4"
) sum areas gained and lost
slf_range_shift_126_table <- terra::expanse(
x = slf_range_shift_126,
unit = "km",
byValue = TRUE
)
slf_range_shift_126_table <- table_range_shift(table.df = slf_range_shift_126_table)
# save as .csv
write_csv(slf_range_shift_126_table, file.path(here::here(), "vignette-outputs", "data-tables", "slf_range_shift_summed_area_table_ssp126_GFDL.csv"))
# convert to kable
slf_range_shift_126_kable <- knitr::kable(x = slf_range_shift_126_table, format = "html", escape = FALSE) %>%
kableExtra::kable_styling(bootstrap_options = "striped", full_width = FALSE)
slf_range_shift_126_kable| Ld_range_shift_type | area_km² | prop_total_area |
|---|---|---|
| remains_unsuitable | 115,684,686 | 75.95% |
| contraction | 3,643,253 | 2.39% |
| expansion | 5,054,355 | 3.32% |
| retained_suitability | 27,927,541 | 18.34% |
# save as .html
kableExtra::save_kable(
slf_range_shift_126_kable,
file = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp126_GFDL.html"),
self_contained = TRUE,
bs_theme = "simplex"
)
# initialize webshot by
# webshot::install_phantomjs()
# convert to pdf
webshot::webshot(
url = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp126_GFDL.html"),
file = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp126_GFDL.jpg"),
zoom = 2
)
# remove .html
file.remove(file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp126_GFDL.html"))Now, I will repeat this process for the other SSP scenarios and the mean of those scenarios (code not shown).
4.5 ssp ssp_mean
classify
slf_binarized_2055_ssp_mean <- terra::rast(x = file.path(mypath, "working_dir", "slf_binarized_summed_2041-2070_ssp_mean_GFDL.asc"))
# rename values
names(slf_binarized_2055_ssp_mean) <- "global_regional_binarized_2055_ssp_mean"
# convert to df
slf_binarized_2055_ssp_mean_df <- terra::as.data.frame(slf_binarized_2055_ssp_mean, xy = TRUE)
# global
# reclassify and write global raster
slf_binarized_2055_ssp_mean_classified <- terra::classify(
x = slf_binarized_2055_ssp_mean,
rcl = classes_2055,
others = strtoi(00000100, base = 2),
filename = file.path(mypath, "working_dir", "slf_range_shift_2041-2070_ssp_mean_GFDL.asc"),
filetype = "AAIGrid",
overwrite = FALSE
) I will run a few error checks to ensure that the binarization process worked.
# take minmax
terra::minmax(slf_binarized_1995_classified) %>%
format(., scientific = FALSE)
terra::minmax(slf_binarized_2055_ssp_mean_classified) %>%
format(., scientific = FALSE)
# all good
# check unique values
unique(values(slf_binarized_1995_classified))
unique(values(slf_binarized_2055_ssp_mean_classified))Mosaic
slf_binarized_1995_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_1981-2010.asc"))
slf_binarized_2055_ssp_mean_classified <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_2041-2070_ssp_mean_GFDL.asc"))
# overlay and combine rasters by summing values
slf_range_shift_ssp_mean <- terra::mosaic(
slf_binarized_1995_classified, slf_binarized_2055_ssp_mean_classified, # rasters
fun = "sum",
filename = file.path(mypath, "working_dir", "slf_range_shift_summed_ssp_mean_GFDL.asc"),
overwrite = FALSE,
wopt = list(
progress = 1,
names = "range_shift_summed"
)
) # progress barPlot
# re-load created raster
slf_range_shift_ssp_mean <- terra::rast(x = file.path(mypath, "working_dir", "slf_range_shift_summed_ssp_mean_GFDL.asc"))
# convert to df
slf_range_shift_ssp_mean_df <- terra::as.data.frame(slf_range_shift_ssp_mean, xy = TRUE) #%>%
# na.omit()
# check for the number of unique values in the raster values column
unique(slf_range_shift_ssp_mean_df$range_shift_summed)## [1] 5 9 10 6
# it seems that the reclassification worked! Lets map it to be sure.
slf_range_shift_ssp_mean_plot <- plot_range_shift(
raster.df = slf_range_shift_ssp_mean_df
) +
# labels
labs(
title = "Projected areas suitable for Lycorma delicatula range expansion",
subtitle = "2041-2070 | mean of ssp126, ssp370 and ssp585 | GFDL-ESM4"
)
slf_range_shift_ssp_mean_plot## Warning: Raster pixels are placed at uneven horizontal intervals and will be shifted
## ℹ Consider using `geom_tile()` instead.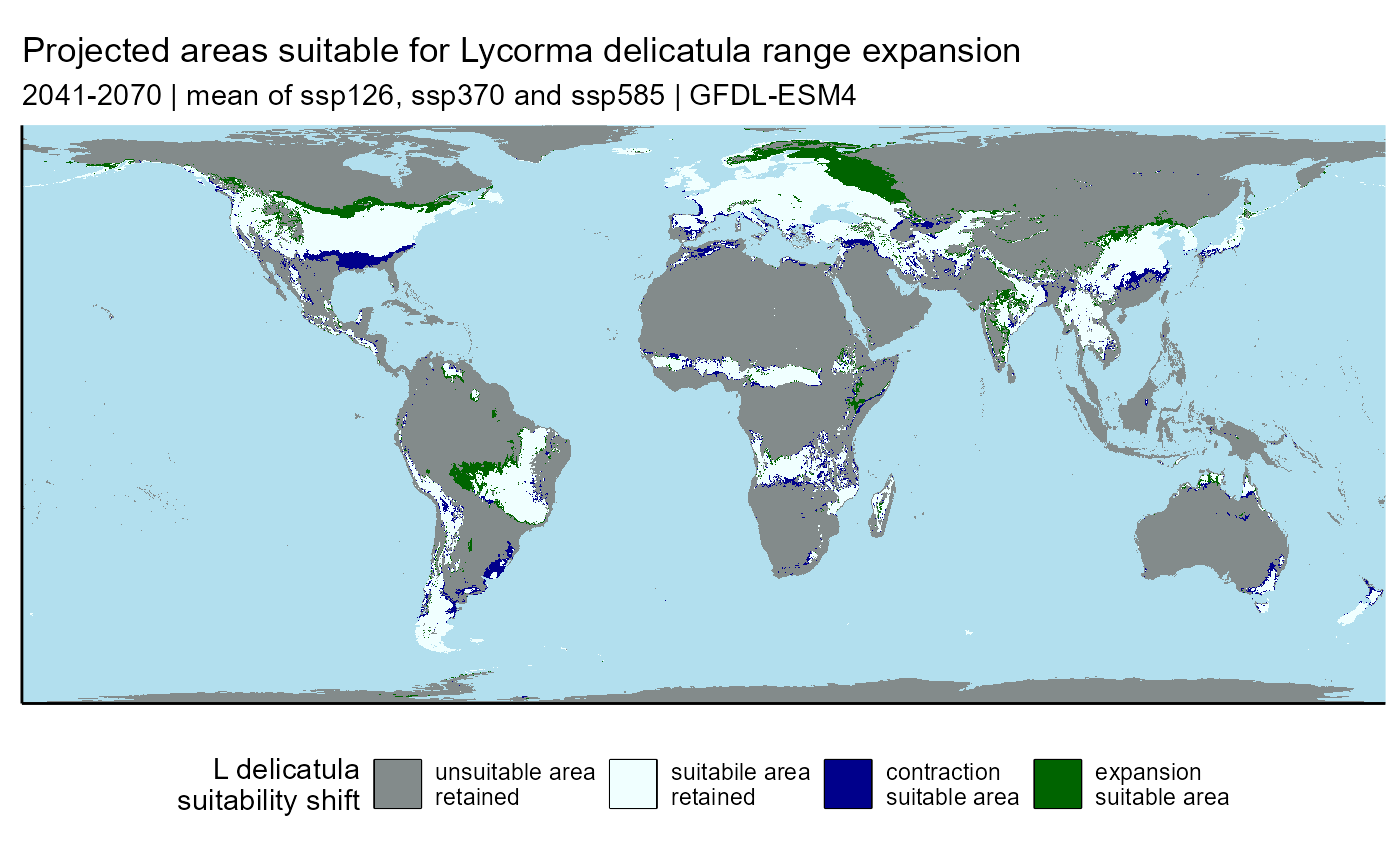
ggsave(
slf_range_shift_ssp_mean_plot,
filename = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_ssp_mean_GFDL.jpg"),
height = 8,
width = 12,
device = jpeg,
dpi = "retina"
)sum areas gained and lost
slf_range_shift_ssp_mean_table <- terra::expanse(
x = slf_range_shift_ssp_mean,
unit = "km",
byValue = TRUE
)
slf_range_shift_ssp_mean_table <- table_range_shift(table.df = slf_range_shift_ssp_mean_table)
# save as .csv
write_csv(slf_range_shift_ssp_mean_table, file.path(here::here(), "vignette-outputs", "data-tables", "slf_range_shift_summed_area_table_ssp_mean_GFDL.csv"))
# convert to kable
slf_range_shift_ssp_mean_kable <- knitr::kable(x = slf_range_shift_ssp_mean_table, format = "html", escape = FALSE) %>%
kableExtra::kable_styling(bootstrap_options = "striped", full_width = FALSE)
slf_range_shift_ssp_mean_kable| Ld_range_shift_type | area_km² | prop_total_area |
|---|---|---|
| remains_unsuitable | 114,450,083 | 75.1% |
| contraction | 4,303,121 | 2.8% |
| expansion | 6,288,957 | 4.1% |
| retained_suitability | 27,267,673 | 17.9% |
# save as .html
kableExtra::save_kable(
slf_range_shift_ssp_mean_kable,
file = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp_mean_GFDL.html"),
self_contained = TRUE,
bs_theme = "simplex"
)
# initialize webshot by
# webshot::install_phantomjs()
# convert to pdf
webshot::webshot(
url = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp_mean_GFDL.html"),
file = file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp_mean_GFDL.jpg"),
zoom = 2
)
# remove .html
file.remove(file.path(here::here(), "vignette-outputs", "figures", "slf_range_shift_summed_area_table_ssp_mean_GFDL.html"))References
Gallien, L., Douzet, R., Pratte, S., Zimmermann, N. E., & Thuiller, W. (2012). Invasive species distribution models – how violating the equilibrium assumption can create new insights. Global Ecology and Biogeography, 21(11), 1126–1136. https://doi.org/10.1111/j.1466-8238.2012.00768.x
Liu, C., White, M., & Newell, G. (2013). Selecting thresholds for the prediction of species occurrence with presence-only data. Journal of Biogeography, 40(4), 778–789. https://doi.org/10.1111/jbi.12058
Liu, C., Newell, G., & White, M. (2016). On the selection of thresholds for predicting species occurrence with presence-only data. Ecology and Evolution, 6(1), 337–348. https://doi.org/10.1002/ece3.1878