
Infer stability of known SLF populations due climate change effects
Samuel M. Owens1
2025-08-05
Source:vignettes/131_create_suitability_xy_plots_SLF.Rmd
131_create_suitability_xy_plots_SLF.RmdOverview
In the last vignette, I created figures to analyze the suitability for SLF establishment at key viticultural regions globally. I will now apply this same framework to analyze the risk of establishment for currently known SLF populations. We will use this to infer the stability of these populations under climate change. We will use the same quadrant plot framework as in the last vignette.
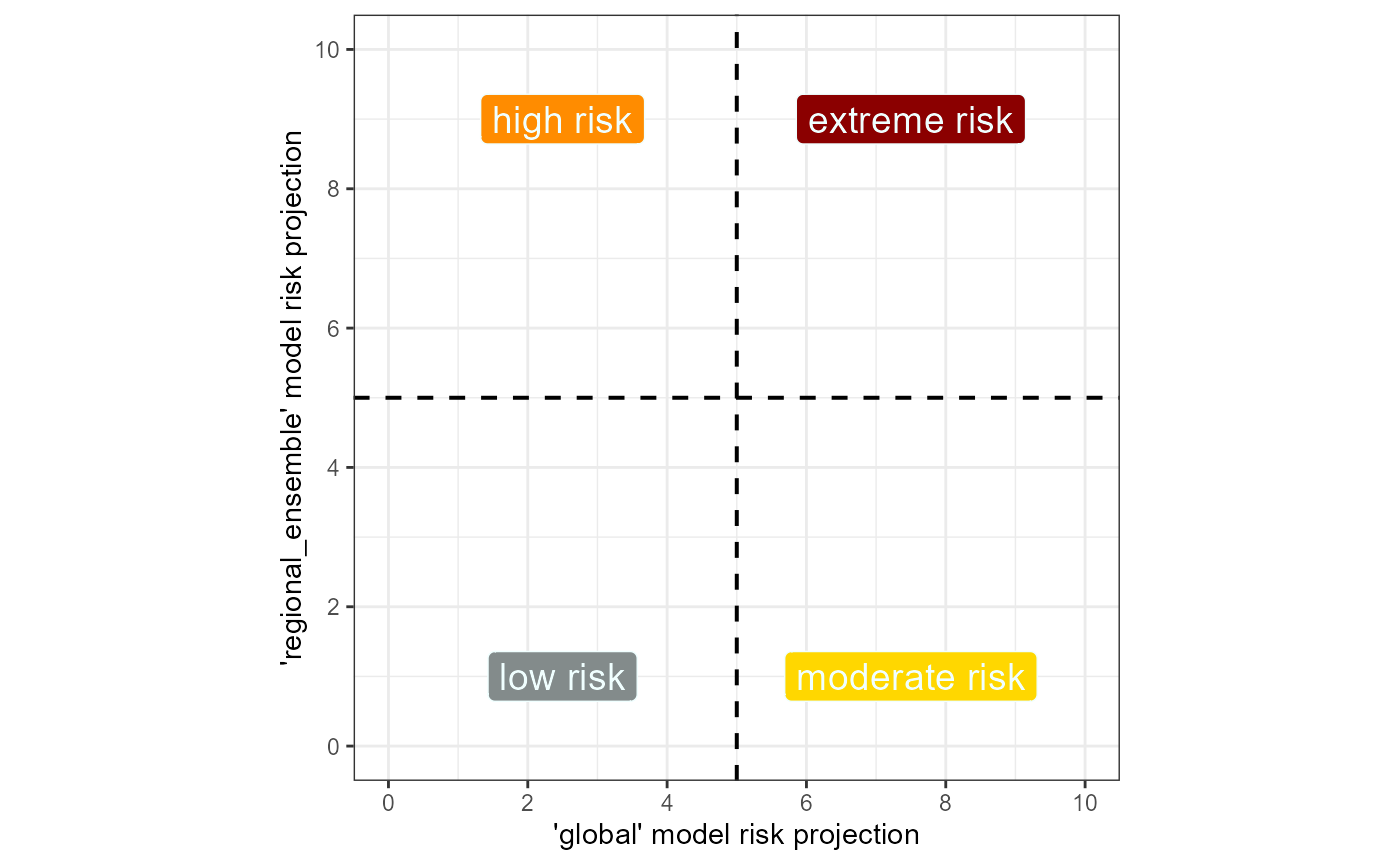 Fig. 1 Example quadrant plot for assessing SLF risk
across two different scales of SDM.
Fig. 1 Example quadrant plot for assessing SLF risk
across two different scales of SDM.
Setup
# general tools
library(tidyverse) #data manipulation
library(here) #making directory pathways easier on different instances
# here() starts at the root folder of this package.
library(devtools)
# spatial data handling
library(terra)
# plot aesthetics
library(scales)
library(patchwork)
library(grid)
library(kableExtra)
library(webshot)
library(webshot2)
library(scari)Note: I will be setting the global options of this
document so that only certain code chunks are rendered in the final
.html file. I will set the eval = FALSE so that none of the
code is re-run (preventing files from being overwritten during knitting)
and will simply overwrite this in chunks with plots.
I will load in some aesthetic objects, including for breaks.
Load in summary files for the global and regional ensemble models, which contain the thresholds.
# summary file to extract thresholds from
# global
summary_global <- read_csv(file = file.path(mypath, "slf_global_v4", "global_summary_all_iterations.csv"))
summary_regional_ensemble <- read_csv(file = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_threshold_values.csv"))load rasters
Finally, I will load in the global and regional ensemble suitability maps. These will be used both for extracting the new xy suitability values and for plotting.
# global
global_1995 <- terra::rast(x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_1981-2010_mean.asc"))
global_2055 <- terra::rast(x = file.path(mypath, "slf_global_v4", "global_pred_suit_clamped_cloglog_globe_2041-2070_GFDL_ssp_averaged.asc"))
# regional_ensemble
# historical
regional_ensemble_1995 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_1981-2010.asc")
)
# CMIP6
## ssp 126
regional_ensemble_2055_126 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp126.asc")
)
# ssp 370
regional_ensemble_2055_370 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp370.asc")
)
# ssp 585
regional_ensemble_2055_585 <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp585.asc")
)
# ssp mean
regional_ensemble_2055_ssp_mean <- terra::rast(
x = file.path(mypath, "slf_regional_ensemble_v2", "ensemble_regional_weighted_mean_globe_2041-2070_GFDL_ssp_averaged.asc")
)tidy slf populations dataset
I will load in the dataset containing SLF populations. I also need to add the “join_cols” to this dataset.
slf_populations <- read_rds(file = file.path(here::here(), "data", "slf_all_coords_final_2025-07-07.rds"))
# round to create join_cols
slf_populations <- slf_populations %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) I will load in one of the xy_suitability files to compare with the slf populations dataset because some points were lost in the predict step, so now the slf populations dataset has more data points than the xy_suitability versions.
# historical
xy_global_1995 <- read_csv(
file = file.path(mypath, "slf_global_v4", "global_slf_all_coords_1981-2010_xy_pred_suit_clamped_cloglog_mean.csv")
)
xy_global_1995 <- xy_global_1995 %>%
# add join columns
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
# rename original cols to keep straight during joins
dplyr::rename(
x_global_1995 = x,
y_global_1995 = y
)
# filter join of populations dataset
slf_populations_join <- dplyr::semi_join(slf_populations, xy_global_1995, by = c("join_col_x", "join_col_y"))
# add ID column
slf_populations_join <- slf_populations_join %>%
dplyr::mutate(ID = row_number()) %>%
dplyr::relocate(ID) %>%
dplyr::select(-c(join_col_x, join_col_y))
# write to file
readr::write_rds(slf_populations_join, file.path(here::here(), "data", "slf_all_coords_final_2025-07-07_tidied.rds"))
# remove object
rm(slf_populations_join)Now that the records have been harmonized between the datasets, I will save to file for use downstream. This will be the main file for predicting slf population suitability.
import and tidy xy suitability
These scatter plots will be based on the suitability for the IVR
points in both the global and regional_ensemble models. I have already
calculated the xy suitability for the global model based on these
points, using the function scari::predict_xy_suitability().
This function will not work for the regional_ensemble because it calls
for a model object, which we did not use to predict the ensemble
suitability. So, I will use terra::extract() to perform
this action.
I will load in the global model datasets and create the regional_ensemble datasets. I will also do some tidying of my datasets for the plots I will create.
# CMIP6
## ssp 126
xy_global_2055_126 <- read_csv(
file = file.path(mypath, "slf_global_v4", "global_slf_all_coords_2041-2070_GFDL_ssp126_xy_pred_suit_clamped_cloglog_mean.csv")
)
## ssp 370
xy_global_2055_370 <- read_csv(
file = file.path(mypath, "slf_global_v4", "global_slf_all_coords_2041-2070_GFDL_ssp370_xy_pred_suit_clamped_cloglog_mean.csv")
)
## ssp 585
xy_global_2055_585 <- read_csv(
file = file.path(mypath, "slf_global_v4", "global_slf_all_coords_2041-2070_GFDL_ssp585_xy_pred_suit_clamped_cloglog_mean.csv")
)
xy_global_2055_126 <- xy_global_2055_126 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::rename(
x_global_2055_126 = x,
y_global_2055_126 = y,
"cloglog_suit_ssp126" = "cloglog_suitability" # rename max column for joining later
) %>%
dplyr::select(-Species)
xy_global_2055_370 <- xy_global_2055_370 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::rename(
x_global_2055_370 = x,
y_global_2055_370 = y,
"cloglog_suit_ssp370" = "cloglog_suitability"
) %>%
dplyr::select(-Species)
xy_global_2055_585 <- xy_global_2055_585 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::rename(
x_global_2055_585 = x,
y_global_2055_585 = y,
"cloglog_suit_ssp585" = "cloglog_suitability"
) %>%
dplyr::select(-Species)take mean of global model ssp scenarios
I will take the mean of the suitability value within each of these predicted suitability datasets to create a single prediction for the three ssp scenarios.
# first join datasets
xy_global_2055_ssp_mean <- xy_global_2055_126 %>%
dplyr::left_join(., xy_global_2055_370, join_by(join_col_x, join_col_y)) %>%
dplyr::left_join(., xy_global_2055_585, join_by(join_col_x, join_col_y)) %>%
# relocate columns
dplyr::relocate(cloglog_suit_ssp126, cloglog_suit_ssp370, cloglog_suit_ssp585) %>%
# take mean of columns
dplyr::mutate(suit_ssp_averaged = rowMeans(.[, 1:3])) %>%
dplyr::select(x_global_2055_126, y_global_2055_126, suit_ssp_averaged) %>%
# rename for saving
dplyr::rename(
x = x_global_2055_126,
y = y_global_2055_126
)retrieve suitability values for regional_ensemble
Now, I will retrieve the suitability values for the regional_ensemble. Instead of returning the coordinates from the map, I will join the coordinates from the original slf_populations dataset so that the coordinates are exact for joining with other datasets. This step is much simpler than in the last vignette because I will be using a simple point-wise prediction method, instead of the buffer method used on the wineries datasets, as these coordinates represent exact population locations, rather than the centroid of a region.
# 1995
xy_regional_ensemble_1995 <- terra::extract(
x = regional_ensemble_1995,
y = dplyr::select(slf_populations, x, y), # points
method = "simple",
xy = FALSE, # dont return coordinates
ID = TRUE
)
# CMIP6
# ssp126
xy_regional_ensemble_2055_126 <- terra::extract(
x = regional_ensemble_2055_126,
y = dplyr::select(slf_populations, x, y), # points
method = "simple",
xy = FALSE, # dont return coordinates
ID = TRUE
)
# ssp370
xy_regional_ensemble_2055_370 <- terra::extract(
x = regional_ensemble_2055_370,
y = dplyr::select(slf_populations, x, y), # points
method = "simple",
xy = FALSE, # dont return coordinates
ID = TRUE
)
# ssp126
xy_regional_ensemble_2055_585 <- terra::extract(
x = regional_ensemble_2055_585,
y = dplyr::select(slf_populations, x, y), # points
method = "simple",
xy = FALSE, # dont return coordinates
ID = TRUE
)
# joining object
slf_coordinates <- slf_populations %>%
dplyr::select(ID, x, y) %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
)
# perform join
xy_regional_ensemble_1995 <- dplyr::left_join(xy_regional_ensemble_1995, slf_coordinates, by = "ID") %>%
# tidy
dplyr::rename(
x_regional_ensemble_1995 = x,
y_regional_ensemble_1995 = y,
"cloglog_suit_hist" = "weighted.mean"
) %>%
dplyr::relocate(ID, x_regional_ensemble_1995, y_regional_ensemble_1995)
# CMIP6
## ssp 126
xy_regional_ensemble_2055_126 <- dplyr::left_join(xy_regional_ensemble_2055_126, slf_coordinates, by = "ID") %>%
# tidy
dplyr::rename(
x_regional_ensemble_2055_126 = x,
y_regional_ensemble_2055_126 = y,
"cloglog_suit_ssp126" = "weighted.mean"
) %>%
dplyr::relocate(ID, x_regional_ensemble_2055_126, y_regional_ensemble_2055_126)
## ssp370
xy_regional_ensemble_2055_370 <- dplyr::left_join(xy_regional_ensemble_2055_370, slf_coordinates, by = "ID") %>%
dplyr::rename(
x_regional_ensemble_2055_370 = x,
y_regional_ensemble_2055_370 = y,
"cloglog_suit_ssp370" = "weighted.mean"
) %>%
dplyr::relocate(ID, x_regional_ensemble_2055_370, y_regional_ensemble_2055_370)
## ssp 585
xy_regional_ensemble_2055_585 <- dplyr::left_join(xy_regional_ensemble_2055_585, slf_coordinates, by = "ID") %>%
dplyr::rename(
x_regional_ensemble_2055_585 = x,
y_regional_ensemble_2055_585 = y,
"cloglog_suit_ssp585" = "weighted.mean"
) %>%
dplyr::relocate(ID, x_regional_ensemble_2055_585, y_regional_ensemble_2055_585)take mean of ssps
# first join datasets
xy_regional_ensemble_2055_ssp_mean <- xy_regional_ensemble_2055_126 %>%
dplyr::left_join(., xy_regional_ensemble_2055_370, join_by(ID, join_col_x, join_col_y)) %>%
dplyr::left_join(., xy_regional_ensemble_2055_585, join_by(ID, join_col_x, join_col_y)) %>%
# relocate columns
dplyr::relocate(cloglog_suit_ssp126, cloglog_suit_ssp370, cloglog_suit_ssp585) %>%
# take mean of columns
dplyr::mutate(suit_ssp_averaged = rowMeans(.[, 1:3])) %>%
dplyr::select(ID, x_regional_ensemble_2055_126, y_regional_ensemble_2055_126, suit_ssp_averaged) %>%
# rename for saving
dplyr::rename(
x = x_regional_ensemble_2055_126,
y = y_regional_ensemble_2055_126
)write to file
Now, I will tidy and save the datasets to .rds for our analysis.
# global model datasets
xy_global_1995 <- xy_global_1995 %>%
# add ID column
cbind(., slf_populations[, 1]) %>%
# rename the column for future joining
dplyr::rename(
x = x_global_1995,
y = y_global_1995,
"xy_global_1995" = "cloglog_suitability"
) %>%
dplyr::relocate(ID) %>%
# remove join columns
dplyr::select(-c(join_col_x, join_col_y))
xy_global_2055_ssp_mean <- xy_global_2055_ssp_mean %>%
# add ID column
cbind(., slf_populations[, 1]) %>%
dplyr::rename(
"xy_global_2055" = "suit_ssp_averaged"
) %>%
dplyr::relocate(ID)
# regional_ensemble datasets
xy_regional_ensemble_1995 <- xy_regional_ensemble_1995 %>%
# revert to original x and y names
dplyr::rename(
x = x_regional_ensemble_1995,
y = y_regional_ensemble_1995,
"xy_regional_ensemble_1995" = "cloglog_suit_hist"
) %>%
# remove join columns
dplyr::select(-c(join_col_x, join_col_y))
xy_regional_ensemble_2055_ssp_mean <- xy_regional_ensemble_2055_ssp_mean %>%
# rename the column for future joining
dplyr::rename("xy_regional_ensemble_2055" = "suit_ssp_averaged")
# save global datasets
readr::write_rds(
xy_global_1995,
file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit.rds")
)
readr::write_rds(
xy_global_2055_ssp_mean,
file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit.rds")
)
# regional_ensemble
readr::write_rds(
xy_regional_ensemble_1995,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit.rds")
)
readr::write_rds(
xy_regional_ensemble_2055_ssp_mean,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit.rds")
)1. Transform xy suitability
I will plot the suitability values in two different ways- I will plot the raw xy suitability and I will transform the data so that the MTSS threshold is the center of the scatter plot. This way, movement across the minimum suitability threshold is more easily visualized. I will transform all 4 vectors of suitability values, 2 per model, in preparation for plotting.
I created the function
scari::rescale_cloglog_suitability() to accomplish this
task. This function uses a vector of exponential transformations for the
specified value of thresh to apply an exponential equation
to the vector of suitability values. It then applies the equation
y = c1 * c2^x + c3 to the vector, where x is the input
suitability values, y is the transformed version of those values, c1 and
c3 are the maximum and its inverse, and c2 is the interpolated value of
the input thresh. The transformed suitability vector is
re-scaled so that thresh is the median (0.5) on a 0-1 scale
and all other values are transformed to fit this scale.
slf_populations <- read_rds(file.path(here::here(), "data", "slf_all_coords_final_2025-07-07_tidied.rds"))
# global
xy_global_1995 <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit.rds"))
xy_global_2055 <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit.rds"))
# regional
xy_regional_ensemble_1995 <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit.rds"))
xy_regional_ensemble_2055 <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit.rds"))
xy_global_1995_rescaled <- scari::rescale_cloglog_suitability(
xy.predicted = xy_global_1995,
thresh = "MTSS",
exponential.file = file.path(here::here(), "data-raw", "threshold_exponential_values.csv"),
summary.file = summary_global,
rescale.name = "xy_global_1995",
rescale.thresholds = TRUE
)
# separate data from thresholds
xy_global_1995_rescaled_thresholds <- xy_global_1995_rescaled[[2]]
xy_global_1995_rescaled <- xy_global_1995_rescaled[[1]]
xy_global_2055_rescaled <- scari::rescale_cloglog_suitability(
xy.predicted = xy_global_2055,
thresh = "MTSS", # the global model only has 1 MTSS thresh
exponential.file = file.path(here::here(), "data-raw", "threshold_exponential_values.csv"),
summary.file = summary_global,
rescale.name = "xy_global_2055",
rescale.thresholds = TRUE
)
xy_global_2055_rescaled_thresholds <- xy_global_2055_rescaled[[2]]
xy_global_2055_rescaled <- xy_global_2055_rescaled[[1]]
xy_regional_ensemble_1995_rescaled <- scari::rescale_cloglog_suitability(
xy.predicted = xy_regional_ensemble_1995,
thresh = "MTSS",
exponential.file = file.path(here::here(), "data-raw", "threshold_exponential_values.csv"),
summary.file = summary_regional_ensemble,
rescale.name = "xy_regional_ensemble_1995",
rescale.thresholds = TRUE
)
xy_regional_ensemble_1995_rescaled_thresholds <- xy_regional_ensemble_1995_rescaled[[2]]
xy_regional_ensemble_1995_rescaled <- xy_regional_ensemble_1995_rescaled[[1]]
xy_regional_ensemble_2055_rescaled <- scari::rescale_cloglog_suitability(
xy.predicted = xy_regional_ensemble_2055,
thresh = "MTSS.CC", # the way the thresholds are calculated for the regional_ensemble model means that the threshold will be slightly different for climate change
exponential.file = file.path(here::here(), "data-raw", "threshold_exponential_values.csv"),
summary.file = summary_regional_ensemble,
rescale.name = "xy_regional_ensemble_2055",
rescale.thresholds = TRUE
)
xy_regional_ensemble_2055_rescaled_thresholds <- xy_regional_ensemble_2055_rescaled[[2]]
xy_regional_ensemble_2055_rescaled <- xy_regional_ensemble_2055_rescaled[[1]]
# global
write_rds(
xy_global_1995_rescaled,
file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit_rescaled.rds")
)
write_rds(
xy_global_2055_rescaled,
file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled.rds")
)
# regional
write_rds(
xy_regional_ensemble_1995_rescaled,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit_rescaled.rds")
)
write_rds(
xy_regional_ensemble_2055_rescaled,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled.rds")
)
# global
write_rds(
xy_global_1995_rescaled_thresholds,
file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit_rescaled_thresholds.rds")
)
write_rds(
xy_global_2055_rescaled_thresholds,
file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled_thresholds.rds")
)
# regional
write_rds(
xy_regional_ensemble_1995_rescaled_thresholds,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit_rescaled_thresholds.rds")
)
write_rds(
xy_regional_ensemble_2055_rescaled_thresholds,
file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled_thresholds.rds")
)2. Plot untransformed suitability values
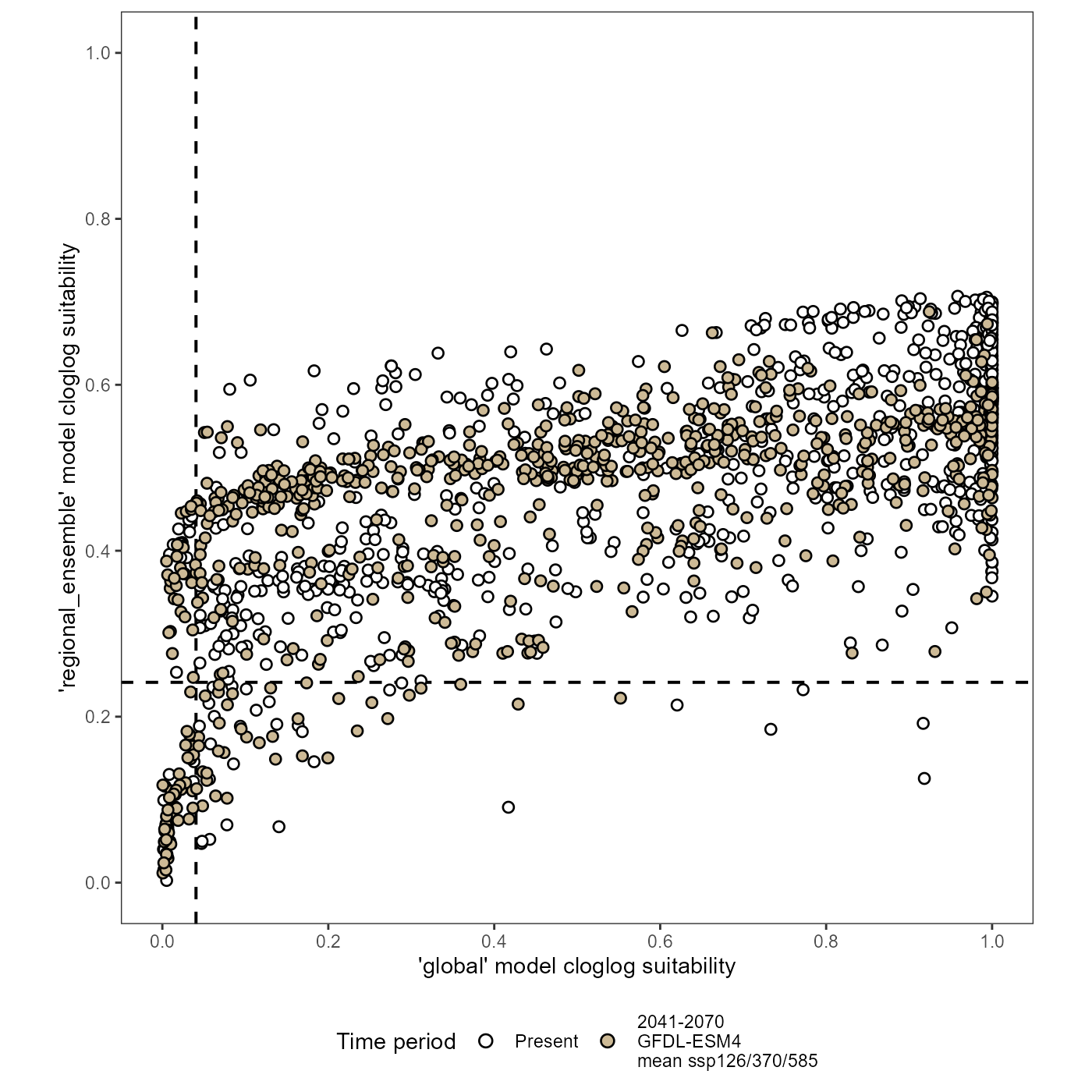
We need a baseline for visualizing the trends in these scatter plots, so I will first plot the un-transformed datasets. I will add join columns again, then join the datasets together for plotting.
# add join columns to all datasets
xy_global_1995 <- xy_global_1995 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # temporarily drop x and y columns
xy_regional_ensemble_1995 <- xy_regional_ensemble_1995 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # temporarily drop x and y columns
xy_global_2055 <- xy_global_2055 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # temporarily drop x and y columns
xy_regional_ensemble_2055 <- xy_regional_ensemble_2055 %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # temporarily drop x and y columns
# join datasets for plotting
xy_joined <- dplyr::full_join(xy_global_1995, xy_regional_ensemble_1995, by = c("ID", "join_col_x", "join_col_y")) %>%
# join CC datasets
dplyr::full_join(., xy_global_2055, by = c("ID", "join_col_x", "join_col_y")) %>%
dplyr::full_join(., xy_regional_ensemble_2055, by = c("ID", "join_col_x", "join_col_y")) %>%
# order
dplyr::relocate(ID, Species, join_col_x, join_col_y, xy_global_1995, xy_global_2055)
# figure annotation title
# "suitability of known Lycorma delicatula populations, projected for climate change"
# plot
(xy_joined_plot <- ggplot(data = xy_joined) +
# threshold lines
# MTSS thresholds
geom_vline(xintercept = as.numeric(summary_global[42, ncol(summary_global)]), linetype = "dashed", linewidth = 0.7) + # global
geom_hline(yintercept = as.numeric(summary_regional_ensemble[6, 4]), linetype = "dashed", linewidth = 0.7) + # regional_ensemble- there are two MTSS thresholds for this model, but the difference is so small that you will never see it on the plot
# historical data
geom_point(
aes(x = xy_global_1995, y = xy_regional_ensemble_1995, shape = "Present"),
size = 2, stroke = 0.7, color = "black", fill = "white"
) +
# GFDL ssp370 data
geom_point(
aes(x = xy_global_2055, y = xy_regional_ensemble_2055, shape = "Future | GFDL-ESM4\nmean of ssp126/370/585"),
size = 2, stroke = 0.7, color = "black", fill = "wheat3"
) +
# axes scaling
scale_x_continuous(name = "'global' model cloglog suitability", limits = c(0, 1), breaks = breaks) +
scale_y_continuous(name = "'regional_ensemble' model cloglog suitability", limits = c(0, 1), breaks = breaks) +
# aesthetics
scale_shape_manual(name = "Time period", values = c(21, 21)) +
guides(shape = guide_legend(nrow = 1, override.aes = list(size = 2.5), reverse = TRUE)) +
theme_bw() +
theme(legend.position = "bottom", panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
coord_fixed(ratio = 1)
)
3. plot transformed suitability values
I will manually change the scale of these values to a 1-10 scale so that this plot of risk is not confused for a measure of suitability from the model.
# global
xy_global_1995_rescaled <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit_rescaled.rds"))
xy_global_2055_rescaled <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled.rds"))
# regional
xy_regional_ensemble_1995_rescaled <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit_rescaled.rds"))
xy_regional_ensemble_2055_rescaled <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled.rds"))
# global
xy_global_1995_rescaled_thresholds <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_1981-2010_xy_pred_suit_rescaled_thresholds.rds"))
xy_global_2055_rescaled_thresholds <- read_rds(file = file.path(here::here(), "data", "global_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled_thresholds.rds"))
# regional
xy_regional_ensemble_1995_rescaled_thresholds <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_1981-2010_xy_pred_suit_rescaled_thresholds.rds"))
xy_regional_ensemble_2055_rescaled_thresholds <- read_rds(file = file.path(here::here(), "data", "regional_ensemble_slf_all_coords_2041-2070_GFDL_ssp_mean_xy_pred_suit_rescaled_thresholds.rds"))
# add join columns to all datasets
xy_global_1995_rescaled <- xy_global_1995_rescaled %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # drop x and y columns
xy_regional_ensemble_1995_rescaled <- xy_regional_ensemble_1995_rescaled %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # drop x and y columns
xy_global_2055_rescaled <- xy_global_2055_rescaled %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # drop x and y columns
xy_regional_ensemble_2055_rescaled <- xy_regional_ensemble_2055_rescaled %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
) %>%
dplyr::select(-c(x, y)) # drop x and y columns
# join datasets for plotting
xy_joined_rescaled <- dplyr::full_join(xy_global_1995_rescaled, xy_regional_ensemble_1995_rescaled, by = c("ID", "join_col_x", "join_col_y")) %>%
# join CC datasets
dplyr::full_join(., xy_global_2055_rescaled, by = c("ID", "join_col_x", "join_col_y")) %>%
dplyr::full_join(., xy_regional_ensemble_2055_rescaled, by = c("ID", "join_col_x", "join_col_y")) %>%
# order
dplyr::relocate(ID, Species, join_col_x, join_col_y, xy_global_1995_rescaled, xy_global_2055_rescaled) %>%
dplyr::select(-c(xy_global_1995, xy_global_2055, xy_regional_ensemble_1995, xy_regional_ensemble_2055))I will need to create a second dataset for the arrow segments indicating change. I will filter out only the segments that cross either threshold and then plot these arrows.
First, I need to isolate the MTSS threshold values.
find points that shift across thresholds
# global
global_MTSS <- as.numeric(xy_global_1995_rescaled_thresholds[2, 2])
# regional ensemble
regional_ensemble_MTSS_1995 <- as.numeric(xy_regional_ensemble_1995_rescaled_thresholds[2, 2])
regional_ensemble_MTSS_2055 <- as.numeric(xy_regional_ensemble_2055_rescaled_thresholds[4, 2])
xy_joined_rescaled_intersects <- xy_joined_rescaled %>%
mutate(
crosses_threshold = dplyr::case_when(
# conditional for starting and ending points that overlap a the threshold
# x-axis
xy_global_1995_rescaled > global_MTSS & xy_global_2055_rescaled < global_MTSS ~ "crosses",
xy_global_1995_rescaled < global_MTSS & xy_global_2055_rescaled > global_MTSS ~ "crosses",
# y-axis
xy_regional_ensemble_1995_rescaled > regional_ensemble_MTSS_2055 & xy_regional_ensemble_2055_rescaled < regional_ensemble_MTSS_2055 ~ "crosses",
xy_regional_ensemble_1995_rescaled < regional_ensemble_MTSS_2055 & xy_regional_ensemble_2055_rescaled > regional_ensemble_MTSS_2055 ~ "crosses",
# else
.default = "does not cross"
)
)
# filter out the crosses
xy_joined_rescaled_intersects <- dplyr::filter(
xy_joined_rescaled_intersects,
crosses_threshold == "crosses"
)plot
Now lets plot the data.
# figure annotation title
# "Risk of Lycorma delicatula establishment in globally important viticultural areas, projected for climate change"
# plot
(xy_joined_rescaled_plot <- ggplot(data = xy_joined_rescaled) +
# threshold lines
# MTSS thresholds
geom_vline(xintercept = global_MTSS, linetype = "dashed", linewidth = 0.7) + # global
geom_hline(yintercept = regional_ensemble_MTSS_1995, linetype = "dashed", linewidth = 0.7) + # regional_ensemble- there are two MTSS thresholds for this model, but the difference is so small that you will never see it on the plot
# arrows indicating change
geom_segment(
data = xy_joined_rescaled_intersects,
aes(
x = xy_global_1995_rescaled,
xend = xy_global_2055_rescaled,
y = xy_regional_ensemble_1995_rescaled,
yend = xy_regional_ensemble_2055_rescaled
),
arrow = grid::arrow(angle = 5.5, type = "closed"), alpha = 0.3, linewidth = 0.25, color = "black"
) +
# historical data
geom_point(
aes(x = xy_global_1995_rescaled, y = xy_regional_ensemble_1995_rescaled, shape = "Present"),
size = 2, stroke = 0.7, color = "black", fill = "white"
) +
# GFDL ssp370 data
geom_point(
aes(x = xy_global_2055_rescaled, y = xy_regional_ensemble_2055_rescaled, shape = "Future | GFDL-ESM4\nmean of ssp126/370/585"),
size = 2, stroke = 0.7, color = "black", fill = "wheat3"
) +
# axes scaling
scale_x_continuous(name = "'global' model risk projection", limits = c(0, 1), breaks = breaks, labels = labels) +
scale_y_continuous(name = "'regional_ensemble' model risk projection", limits = c(0, 1), breaks = breaks, labels = labels) +
# quadrant labels
# extreme risk, top right, quad4
geom_label(aes(x = 0.75, y = 0.9, label = "extreme risk"), fill = "darkred", color = "azure", size = 5) +
# high risk, top left, quad3
geom_label(aes(x = 0.25, y = 0.9, label = "high risk"), fill = "darkorange", color = "azure", size = 5) +
# moderate risk, bottom right, quad2
geom_label(aes(x = 0.75, y = 0.1, label = "moderate risk"), fill = "gold", color = "azure", size = 5) +
# low risk, bottom left, quad1
geom_label(aes(x = 0.25, y = 0.1, label = "low risk"), fill = "azure4", color = "azure", size = 5) +
# aesthetics
scale_shape_manual(name = "Time period", values = c(21, 21)) +
guides(shape = guide_legend(nrow = 1, override.aes = list(size = 2.5), reverse = TRUE)) +
theme_bw() +
theme(legend.position = "bottom", panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
coord_fixed(ratio = 1)
)## Warning in geom_label(aes(x = 0.75, y = 0.9, label = "extreme risk"), fill = "darkred", : All aesthetics have length 1, but the data has 1277 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.## Warning in geom_label(aes(x = 0.25, y = 0.9, label = "high risk"), fill = "darkorange", : All aesthetics have length 1, but the data has 1277 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.## Warning in geom_label(aes(x = 0.75, y = 0.1, label = "moderate risk"), fill = "gold", : All aesthetics have length 1, but the data has 1277 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.## Warning in geom_label(aes(x = 0.25, y = 0.1, label = "low risk"), fill = "azure4", : All aesthetics have length 1, but the data has 1277 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.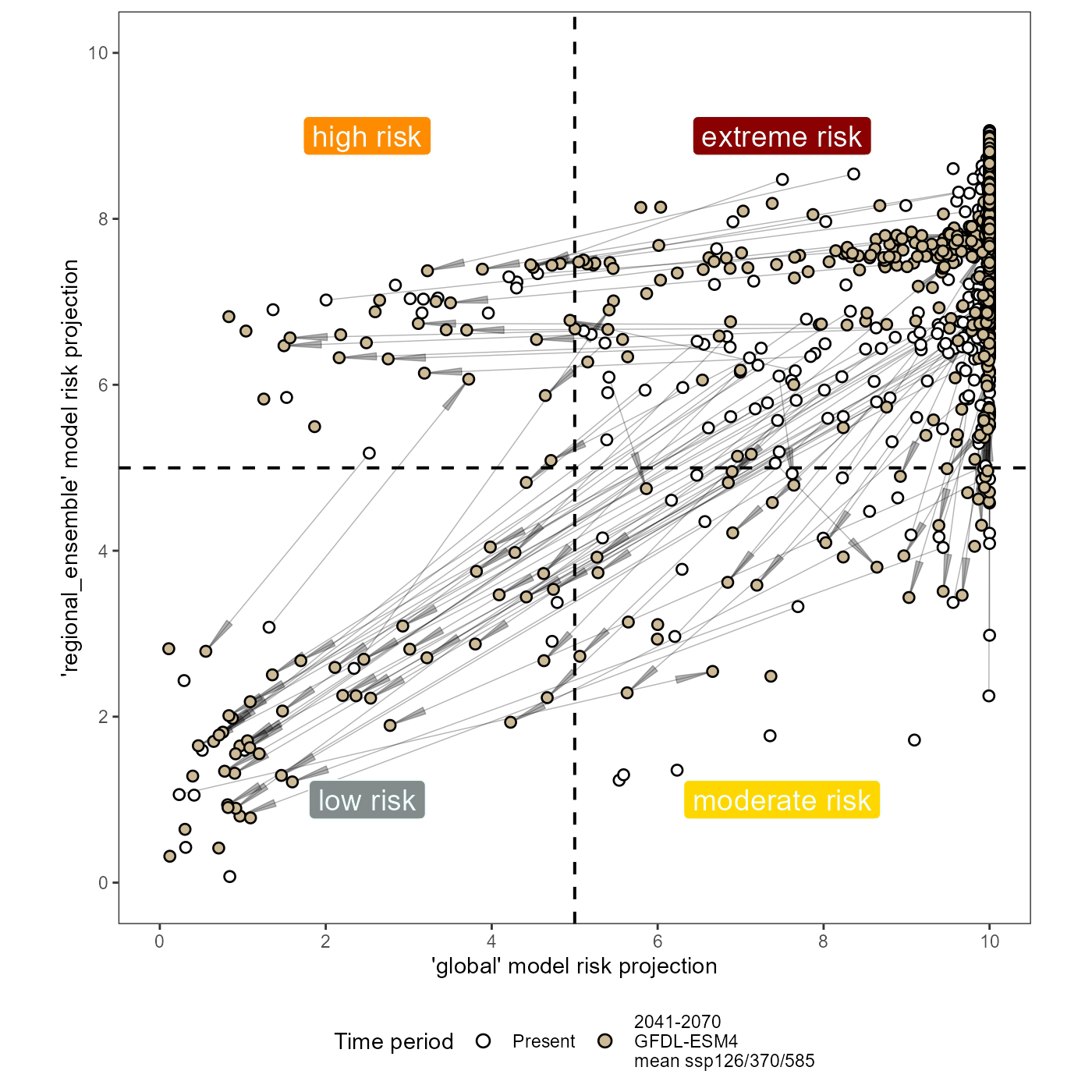
ggsave(
xy_joined_rescaled_plot,
filename = file.path(
here::here(), "vignette-outputs", "figures", "slf_risk_plot.jpg"
),
height = 8,
width = 8,
device = jpeg,
dpi = "retina"
)
# also save to rds
write_rds(
xy_joined_rescaled_plot,
file = file.path(here::here(), "vignette-outputs", "figures", "figures-rds", "slf_risk_plot.rds")
)4. Create summary table of transformed plot
I will now create a summary table to explain the rescaled plots from
step 4. The table will depict the quadrant placement of the point in the
quadrant plot, both before and after climate change. From this, I will
calculate the total number of movements into and out of each quadrant. I
will apply the internal function
scari::calculate_risk_quadrant().
I will create a summary table of the quadrant placement (and thus the
level of risk) for each point in the IVR_locations dataset. I will use
calculate_risk_quadrant() to accomplish this.
# edit slf_populations first
slf_populations <- slf_populations %>%
dplyr::mutate(
join_col_x = round(x, 5),
join_col_y = round(y, 4) # rounding to the 1000s (1km) place to prevent overly sensitive exclusions for UTM data
)
# create dataset and tidy
slf_populations_joined <- dplyr::left_join(slf_populations, xy_joined_rescaled, by = c("ID", "join_col_x", "join_col_y")) %>%
relocate(ID, x, y) %>%
# remove join cols
dplyr::select(-c(join_col_x, join_col_y, Species))
# calculate risk quadrants
slf_populations_risk <- slf_populations_joined %>%
dplyr::mutate(
risk_1995 = scari::calculate_risk_quadrant(
suit.x = slf_populations_joined$xy_global_1995_rescaled,
suit.y = slf_populations_joined$xy_regional_ensemble_1995_rescaled,
thresh.x = global_MTSS, # this threshold remains the same
thresh.y = regional_ensemble_MTSS_1995
),
risk_2055 = scari::calculate_risk_quadrant(
suit.x = slf_populations_joined$xy_global_2055_rescaled,
suit.y = slf_populations_joined$xy_regional_ensemble_2055_rescaled,
thresh.x = global_MTSS,
thresh.y = regional_ensemble_MTSS_2055
),
risk_shift = stringr::str_c(risk_1995, risk_2055, sep = "-")
)
# factor levels for risk categories
risk_levels <- c("extreme", "high", "moderate", "low")
# number of records
n_records <- nrow(slf_populations_risk)
slf_risk_table <- slf_populations_risk %>%
# create counts and make into acrostic table
dplyr::group_by(risk_1995, risk_2055) %>%
dplyr::summarize(count = n()) %>%
pivot_wider(names_from = risk_2055, values_from = count) %>%
# tidy
ungroup()## `summarise()` has grouped output by 'risk_1995'. You can override using the
## `.groups` argument.
# add columns that do not exist
if(!'extreme' %in% names(slf_risk_table)) slf_risk_table <- slf_risk_table %>% tibble::add_column(extreme = 0)
if(!'high' %in% names(slf_risk_table)) slf_risk_table <- slf_risk_table %>% tibble::add_column(high = 0)
if(!'moderate' %in% names(slf_risk_table)) slf_risk_table <- slf_risk_table %>% tibble::add_column(moderate = 0)
if(!'low' %in% names(slf_risk_table)) slf_risk_table <- slf_risk_table %>% tibble::add_column(low = 0)
# ensure all combinations of risk exist
if(!'extreme' %in% slf_risk_table$risk_1995) slf_risk_table <- slf_risk_table %>% tibble::add_row(risk_1995 = "extreme", extreme = 0, high = 0, moderate = 0, low = 0)
if(!'high' %in% slf_risk_table$risk_1995) slf_risk_table <- slf_risk_table %>% tibble::add_row(risk_1995 = "high", extreme = 0, high = 0, moderate = 0, low = 0)
if(!'moderate' %in% slf_risk_table$risk_1995) slf_risk_table <- slf_risk_table %>% tibble::add_row(risk_1995 = "moderate", extreme = 0, high = 0, moderate = 0, low = 0)
if(!'low' %in% slf_risk_table$risk_1995) slf_risk_table <- slf_risk_table %>% tibble::add_row(risk_1995 = "low", extreme = 0, high = 0, moderate = 0, low = 0)
slf_risk_table <- slf_risk_table %>%
dplyr::rename("rows_1995_cols_2055" = "risk_1995") %>%
dplyr::relocate("rows_1995_cols_2055", "extreme", "high", "moderate") %>%
dplyr::arrange(factor(.$rows_1995_cols_2055, levels = risk_levels)) %>%
# replace missing categories with 0
replace(is.na(.), 0)
# tidy
slf_risk_table <- slf_risk_table %>%
# add totals column
tibble::add_column("total_present" = rowSums(.[, 2:5])) %>%
# add row totals
tibble::add_row(rows_1995_cols_2055 = "total_2055", extreme = colSums(dplyr::select(., 2)), high = colSums(dplyr::select(., 3)), moderate = colSums(dplyr::select(., 4)), low = colSums(dplyr::select(., 5)), total_present = n_records) 5. global risk shift vs regional risk shift
I will create a table to sum the number of points in three different groups. My goal is to understand how the regional model adds resolution to our calculation of risk. So, I will look at how
I will sum the number of points that are suitable in the global model only, unsuitable in the global model only, and unsuitable in the global model / suitable in the regional model. I will repeat this operation for both time periods.
global_regional_risk_shift <- tibble(
time_period = c(1995, 1995, 1995, 2055, 2055, 2055),
quadrants = c("quad4_quad2", "quad3_quad1", "quad3", "quad4_quad2", "quad3_quad1", "quad3"),
risk = c("extreme_moderate", "high_low", "high", "extreme_moderate", "high_low", "high"),
model_suit = c("global_suit", "global_unsuit", "global_unsuit_regional_suit", "global_suit", "global_unsuit", "global_unsuit_regional_suit"),
slf_population_count = c(
# global suitable 1995
sum(slf_populations_joined$xy_global_1995_rescaled >= global_MTSS),
# global unsuitable 1995
sum(slf_populations_joined$xy_global_1995_rescaled < global_MTSS),
# global unsuitable and regional suitable 1995
sum(slf_populations_joined$xy_global_1995_rescaled < global_MTSS & slf_populations_joined$xy_regional_ensemble_1995_rescaled >= regional_ensemble_MTSS_1995),
# global suitable 2055
sum(slf_populations_joined$xy_global_2055_rescaled >= global_MTSS),
# global unsuitable 2055
sum(slf_populations_joined$xy_global_2055_rescaled < global_MTSS),
# global unsuitable and regional suitable 2055
sum(slf_populations_joined$xy_global_2055_rescaled < global_MTSS & slf_populations_joined$xy_regional_ensemble_2055_rescaled >= regional_ensemble_MTSS_2055)
)
)
# total # slf populations
total_slf <- sum(global_regional_risk_shift[1:2, 5])
global_regional_risk_shift <- mutate(
global_regional_risk_shift,
slf_population_prop = slf_population_count / total_slf
)
# calculate % of unsuit (quad3 and quad 1) that are are in quad3
quad3_risk_prop <- tibble(
time_period = c("quad3_1995", "quad3_2055"),
prop_total_unsuit_in_quad3 = c(
scales::label_percent(accuracy = 0.01) (abs(as.numeric((global_regional_risk_shift[3, 5]) / global_regional_risk_shift[2, 5]))),
scales::label_percent(accuracy = 0.01) (abs(as.numeric((global_regional_risk_shift[6, 5]) / global_regional_risk_shift[5, 5])))
)
)With this analysis, I found that currently, only about 2.9% of known (rarefied) SLF populations are low risk according to the global model. However, 1.1% of the total populations are specifically in quadrant 3 (high risk). This means that the global model alone would label 22 of the 769 slf populations as low risk, when in actuality 9 or about 40.9% of these are at high risk of persisting (above the MTSS threshold) when we spatially segment the presence data into an ensemble of regional-scale models. After climate change, 78 of the 1063 populations (10%) would be unsuitable if the global model alone were used to describe the risk of SLF. However, 22 or 28% of these unsuitable populations are still suitable in regional_scale models and thus would be missed by an analysis of risk using only a global-scale model.
This means that our regional-scale ensemble is adding resolution and nuance to our estimation of risk for SLF establishment.
# add %
global_regional_risk_shift <- mutate(global_regional_risk_shift, slf_population_prop = scales::label_percent(accuracy = 0.01) (slf_population_prop))
# make kable
global_regional_risk_shift <- kable(global_regional_risk_shift, "html", escape = FALSE) %>%
kableExtra::kable_styling(bootstrap_options = "striped", full_width = FALSE) %>%
kableExtra::add_header_above(., header = c("SLF risk plot quadrant proportions" = 6), bold = TRUE)
# save as .html
kableExtra::save_kable(
global_regional_risk_shift,
file = file.path(here::here(), "vignette-outputs", "figures", "slf_risk_plot_quadrant_props.html"),
self_contained = TRUE
)
# initialize webshot by
# webshot::install_phantomjs()
# convert to pdf
webshot::webshot(
url = file.path(here::here(), "vignette-outputs", "figures", "slf_risk_plot_quadrant_props.html"),
file = file.path(here::here(), "vignette-outputs", "figures", "slf_risk_plot_quadrant_props.jpg"),
zoom = 2
)
# rm html
file.remove(file.path(here::here(), "vignette-outputs", "figures", "slf_risk_plot_quadrant_props.html"))References
Gallien, L., Douzet, R., Pratte, S., Zimmermann, N. E., & Thuiller, W. (2012). Invasive species distribution models – how violating the equilibrium assumption can create new insights. Global Ecology and Biogeography, 21(11), 1126–1136. https://doi.org/10.1111/j.1466-8238.2012.00768.x
Smith, T. 2021, August 11. Evaluating Invasion Stage with SDMs - plantarum.ca. https://plantarum.ca/2021/08/11/invasion-stage/.