Variable importance
regional_native_var_imp <- read.csv(file = file.path(mypath, "slf_regional_native_v3", "regional_native_variable_importance.csv"))
regional_invaded_var_imp <- read.csv(file = file.path(mypath, "slf_regional_invaded_v7", "regional_invaded_variable_importance.csv"))
regional_invaded_asian_var_imp <- read.csv(file = file.path(mypath, "slf_regional_invaded_asian_v2", "regional_invaded_asian_variable_importance.csv"))
regional_native_var_imp_plot <- SDMtune::plotVarImp(
df = regional_native_var_imp
) %>%
ggplot_build()
# change groups
regional_native_var_imp_plot[["data"]][[1]][["x"]] <- c(1, 4, 2, 3)
regional_invaded_var_imp_plot <- SDMtune::plotVarImp(
df = regional_invaded_var_imp
) %>%
ggplot_build()
# change groups
regional_invaded_var_imp_plot[["data"]][[1]][["x"]] <- c(1, 2, 3, 4)
regional_invaded_asian_var_imp_plot <- SDMtune::plotVarImp(
df = regional_invaded_asian_var_imp
) %>%
ggplot_build()
# change groups
regional_invaded_asian_var_imp_plot[["data"]][[1]][["x"]] <- c(1, 2, 4, 3)
var_imp_ensemble <- ggplot() +
# native model data
geom_col(data = regional_native_var_imp_plot$data[[1]], aes(x = x + 0.2, y = y, fill = "Rn (native)"), color = "black", width = 0.2) +
# invaded model data
geom_col(data = regional_invaded_var_imp_plot$data[[1]], aes(x = x, y = y, fill = "Ri.NAmerica"), color = "black", width = 0.2) +
# invaded_asian model data
geom_col(data = regional_invaded_asian_var_imp_plot$data[[1]], aes(x = x - 0.2, y = y, fill = "Ri.Asia"), color = "black", width = 0.2) +
labs(
title = "Variable Importance for 'regional_ensemble' models",
x = "",
y = "Permutation importance"
) +
scale_x_continuous(
breaks = c(1, 2, 3, 4),
labels = c("bio 2", "bio 12", "bio 11", "bio 15")
) +
scale_y_continuous(labels = scales::percent) +
# aes
theme_bw() +
scale_fill_manual(
name = "model",
values = ensemble_colors,
aesthetics = "fill"
) +
theme(legend.position = "bottom") +
coord_flip()
ggsave(
var_imp_ensemble,
filename = file.path(
here::here(), "vignette-outputs", "figures", "Variable_importance_regional_ensemble.jpg"
),
height = 8,
width = 8,
device = jpeg,
dpi = "retina"
)
global_var_imp <- read.csv(file = file.path(mypath, "slf_global_v3", "global_variable_importance.csv"))
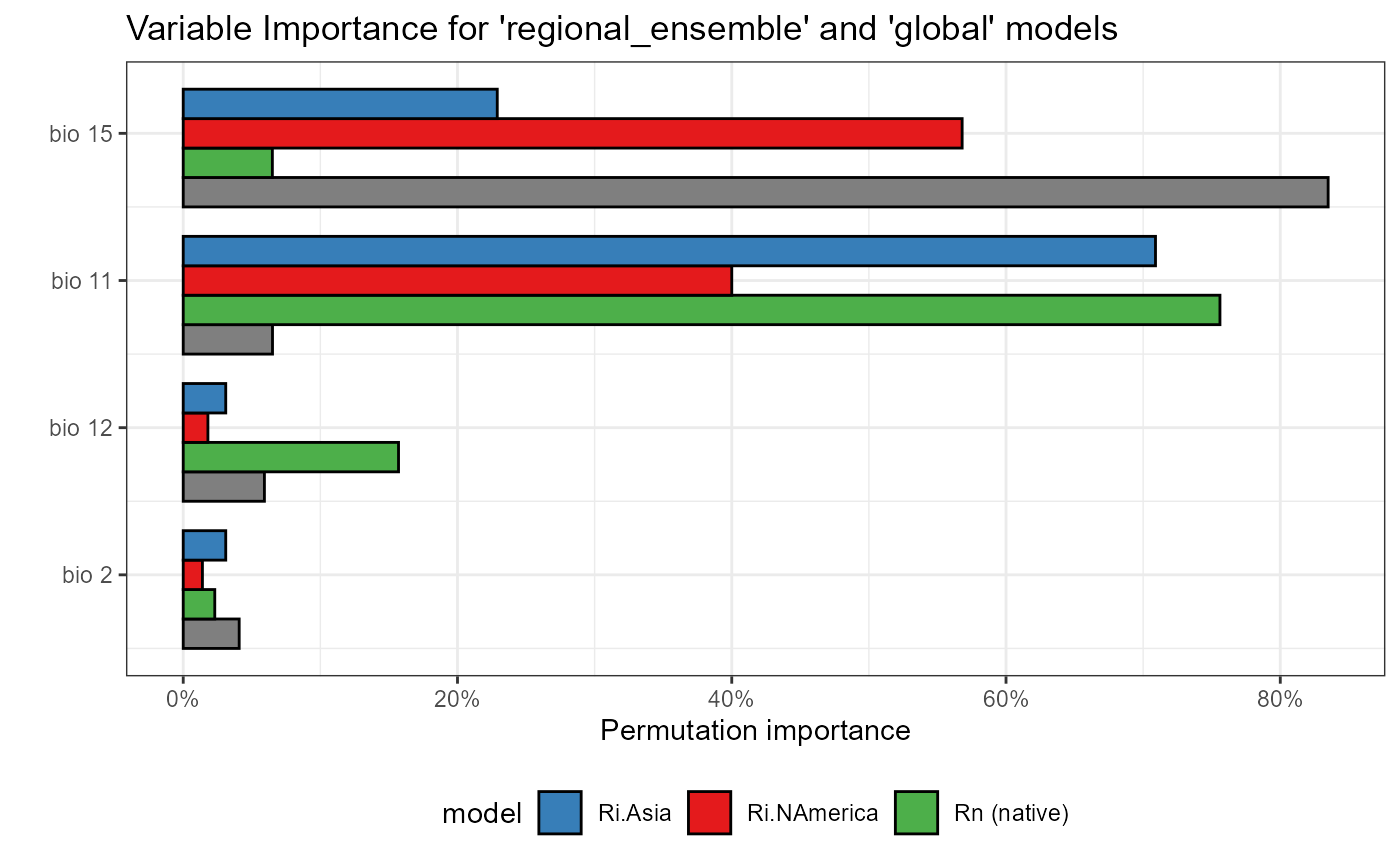
var_imp_ensemble_global <- ggplot() +
# native model data
geom_col(data = global_var_imp_plot$data[[1]], aes(x = x - 0.4, y = y, fill = "global"), color = "black", width = 0.2) +
# native model data
geom_col(data = regional_native_var_imp_plot$data[[1]], aes(x = x - 0.2, y = y, fill = "Rn (native)"), color = "black", width = 0.2) +
# invaded model data
geom_col(data = regional_invaded_var_imp_plot$data[[1]], aes(x = x, y = y, fill = "Ri.NAmerica"), color = "black", width = 0.2) +
# invaded_asian model data
geom_col(data = regional_invaded_asian_var_imp_plot$data[[1]], aes(x = x + 0.2, y = y, fill = "Ri.Asia"), color = "black", width = 0.2) +
labs(
title = "Variable Importance for 'regional_ensemble' and 'global' models",
x = "",
y = "Permutation importance"
) +
scale_x_continuous(
breaks = c(1, 2, 3, 4),
labels = c("bio 2", "bio 12", "bio 11", "bio 15")
) +
scale_y_continuous(labels = scales::percent) +
# aes
theme_bw() +
scale_fill_manual(
name = "model",
values = ensemble_colors,
aesthetics = "fill"
) +
theme(legend.position = "bottom") +
coord_flip()
var_imp_ensemble_global
ggsave(
var_imp_ensemble_global,
filename = file.path(
here::here(), "vignette-outputs", "figures", "Variable_importance_regional_ensemble_global.jpg"
),
height = 8,
width = 8,
device = jpeg,
dpi = "retina"
)

